Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119471 |
| Name | oriT_pKP18-31-3 |
| Organism | Klebsiella quasipneumoniae strain KP18-31 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MN661404 (47681..47730 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKP18-31-3
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file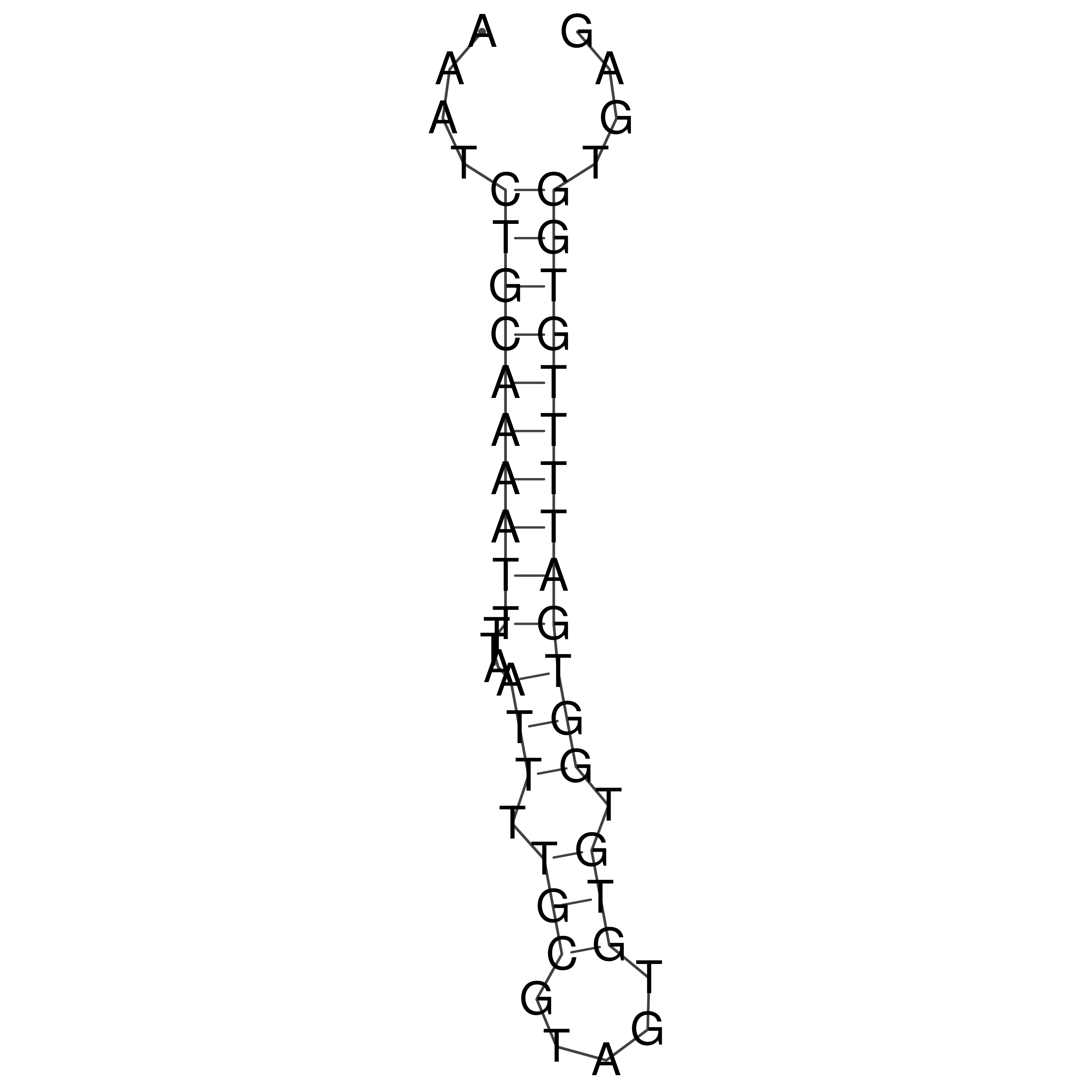
Relaxase
| ID | 12298 | GenBank | WP_043907038 |
| Name | traI_H5G48_RS00500_pKP18-31-3 |
UniProt ID | A0A0S1TKY9 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190458.19 Da Isoelectric Point: 6.0263
>WP_043907038.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSLSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSLSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14433 | GenBank | WP_228723713 |
| Name | traD_H5G48_RS00495_pKP18-31-3 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85998.02 Da Isoelectric Point: 5.1787
>WP_228723713.1 type IV conjugative transfer system coupling protein TraD [Klebsiella quasipneumoniae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTTKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTTKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 47123..76213
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H5G48_RS00285 | 43142..43498 | + | 357 | WP_019706019 | hypothetical protein | - |
| H5G48_RS00290 | 43559..43771 | + | 213 | WP_019706020 | hypothetical protein | - |
| H5G48_RS00295 | 43782..44006 | + | 225 | WP_014343499 | hypothetical protein | - |
| H5G48_RS00300 | 44087..44407 | + | 321 | WP_015065521 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| H5G48_RS00305 | 44397..44675 | + | 279 | WP_020801586 | helix-turn-helix transcriptional regulator | - |
| H5G48_RS00310 | 44676..45077 | + | 402 | WP_015065522 | type II toxin-antitoxin system HigA family antitoxin | - |
| H5G48_RS00315 | 45907..46728 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| H5G48_RS00320 | 46761..47090 | + | 330 | WP_053390269 | DUF5983 family protein | - |
| H5G48_RS00325 | 47123..47608 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| H5G48_RS00330 | 48043..48435 | + | 393 | WP_032441878 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| H5G48_RS00335 | 48651..49337 | + | 687 | WP_071561590 | transcriptional regulator TraJ family protein | - |
| H5G48_RS00340 | 49421..49792 | + | 372 | WP_181726206 | TraY domain-containing protein | - |
| H5G48_RS00345 | 49846..50214 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| H5G48_RS00350 | 50228..50533 | + | 306 | WP_080923122 | type IV conjugative transfer system protein TraL | traL |
| H5G48_RS00355 | 50553..51119 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| H5G48_RS00360 | 51106..51846 | + | 741 | WP_080923121 | type-F conjugative transfer system secretin TraK | traK |
| H5G48_RS00365 | 51846..53270 | + | 1425 | WP_181726207 | F-type conjugal transfer pilus assembly protein TraB | traB |
| H5G48_RS00370 | 53384..53968 | + | 585 | WP_032441881 | type IV conjugative transfer system lipoprotein TraV | traV |
| H5G48_RS00375 | 54100..54510 | + | 411 | WP_004152499 | hypothetical protein | - |
| H5G48_RS00380 | 54616..54834 | + | 219 | WP_004195235 | hypothetical protein | - |
| H5G48_RS00385 | 54835..55146 | + | 312 | WP_004195240 | hypothetical protein | - |
| H5G48_RS00390 | 55213..55617 | + | 405 | WP_004197817 | hypothetical protein | - |
| H5G48_RS00395 | 55994..56392 | + | 399 | WP_014343490 | hypothetical protein | - |
| H5G48_RS00400 | 56464..59103 | + | 2640 | WP_013214024 | type IV secretion system protein TraC | virb4 |
| H5G48_RS00405 | 59103..59492 | + | 390 | WP_013214025 | type-F conjugative transfer system protein TrbI | - |
| H5G48_RS00410 | 59492..60127 | + | 636 | WP_013214026 | type-F conjugative transfer system protein TraW | traW |
| H5G48_RS00415 | 60162..60563 | + | 402 | WP_004194979 | hypothetical protein | - |
| H5G48_RS00420 | 60560..61549 | + | 990 | WP_013214027 | conjugal transfer pilus assembly protein TraU | traU |
| H5G48_RS00425 | 61562..62200 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| H5G48_RS00430 | 62259..64214 | + | 1956 | WP_181726202 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| H5G48_RS00435 | 64246..64500 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| H5G48_RS00440 | 64478..64726 | + | 249 | WP_013214029 | hypothetical protein | - |
| H5G48_RS00445 | 64739..65065 | + | 327 | WP_013214030 | hypothetical protein | - |
| H5G48_RS00450 | 65086..65838 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| H5G48_RS00455 | 65849..66088 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| H5G48_RS00460 | 66060..66617 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| H5G48_RS00465 | 66663..67106 | + | 444 | WP_013214032 | F-type conjugal transfer protein TrbF | - |
| H5G48_RS00470 | 67084..68463 | + | 1380 | WP_074160414 | conjugal transfer pilus assembly protein TraH | traH |
| H5G48_RS00475 | 68463..71285 | + | 2823 | WP_013214034 | conjugal transfer mating-pair stabilization protein TraG | traG |
| H5G48_RS00480 | 71308..71910 | + | 603 | WP_013214035 | conjugal transfer protein TraS | - |
| H5G48_RS00485 | 72161..72892 | + | 732 | WP_013214036 | conjugal transfer complement resistance protein TraT | - |
| H5G48_RS00490 | 73085..73774 | + | 690 | WP_014343485 | hypothetical protein | - |
| H5G48_RS00495 | 73901..76213 | + | 2313 | WP_228723713 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 19901 | GenBank | NZ_MN661404 |
| Plasmid name | pKP18-31-3 | Incompatibility group | IncFII |
| Plasmid size | 107000 bp | Coordinate of oriT [Strand] | 47681..47730 [-] |
| Host baterium | Klebsiella quasipneumoniae strain KP18-31 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsR, arsD, arsA, arsB, arsC |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |