Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119253 |
| Name | oriT_pET-ATCC-159-1 |
| Organism | Edwardsiella tarda ATCC 15947 = NBRC 105688 strain ATCC 15947 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP084508 (124871..124906 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAACT..AGTTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pET-ATCC-159-1
AAATCAGCAAAACTTTAGTTTTGCGTAGTGTGTGGT
AAATCAGCAAAACTTTAGTTTTGCGTAGTGTGTGGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file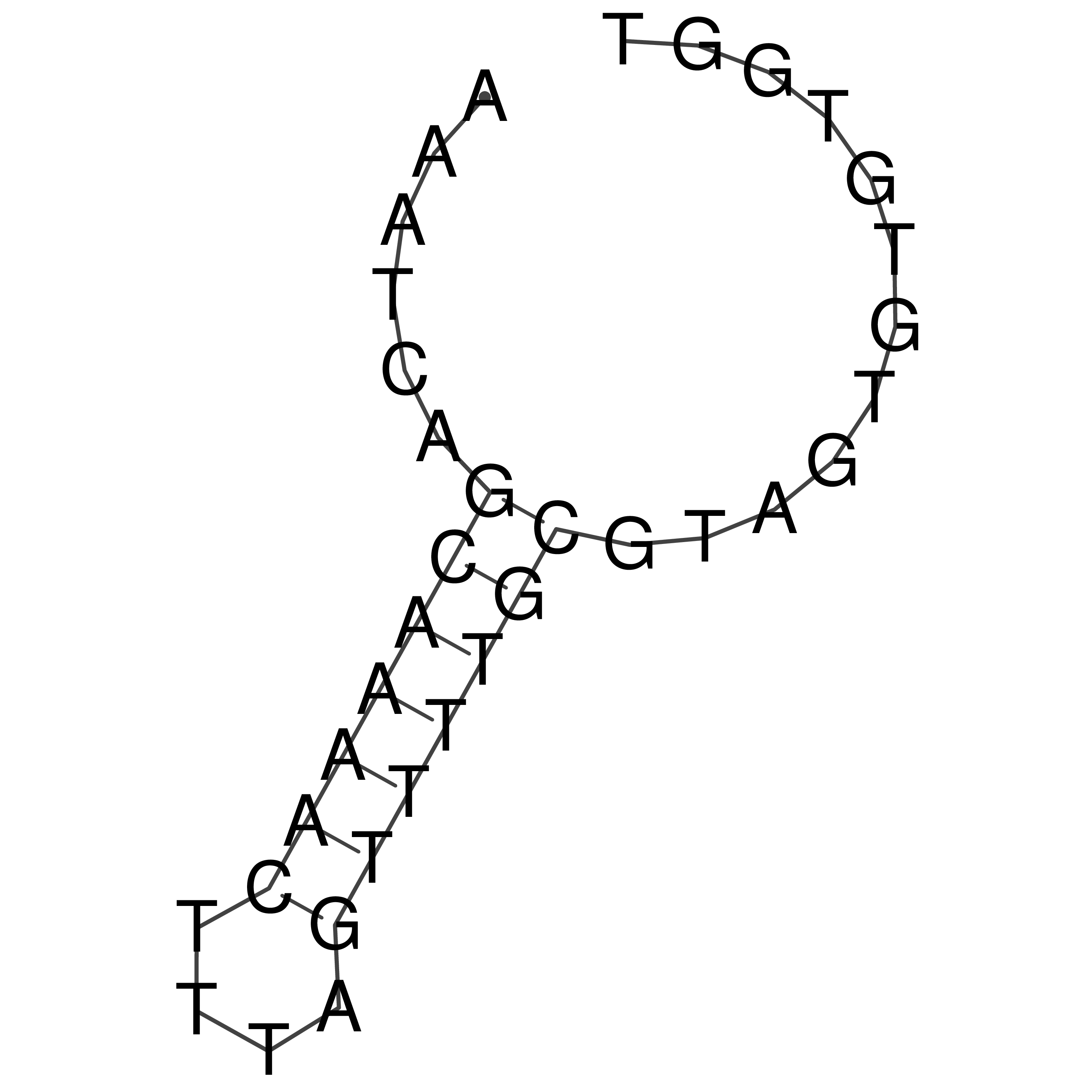
Relaxase
| ID | 12178 | GenBank | WP_035599333 |
| Name | traI_DCL27_RS17400_pET-ATCC-159-1 |
UniProt ID | _ |
| Length | 1745 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1745 a.a. Molecular weight: 188197.94 Da Isoelectric Point: 6.6461
>WP_035599333.1 conjugative transfer relaxase/helicase TraI [Edwardsiella tarda]
MMSFSQVKSASNASGYYTDKDNYYVLGSMDDHWYGEGAKQLGLSGAVDKAVFTQLLHGRLPDGADLSRLE
NGVNKHRPGYDLTFSAPKSVSMMAMLGGDRRLIDAHNRAVDVALRQAEALASTRSMRDGVSETVLTGNLI
IARFNHDTSRDQDPQLHTHSVVLNATRNGDKWQALSSDTVGKTGFSENILANRIAIGKIYNQALRSEVEA
MGYQVEVVGKHGLWEMKNVPVEAFSHRSQAIQEAVGADASLKSRDVAALDTRKAKTAIDPTEKMAEWMAT
LKSTGFDIQGYREAAAQRVTQGQVPAPADPAAVDMAKTVRQAISQLSERKVQFTYSDMLAATVSLLPAND
GVIARARAGIDHAIEQQQLIPLDREKGVFTSDIHVLDELSVRTLGQSLGEQGHVTVGAMASSASRHDYSP
LAQQVMTERPPLAMLASPGGAGVQRQRVSELVQATQALGREVQILVSDAKSQRFLQQDAELGQAAIFPQR
VLNDGMAFTPNSTLIVTEGEKLTLKETVTLLDGALRNNVQLLILDTAQRQATGNALSVLRESGIDTLHYA
QTAPVETQILSEPDKPSRFARLADDYVAARQAGDPVVAQVSGPTDQQHLTRAIRSAMKTQGALGQNDTTL
MALEPVWLDSKNRLSRDTYRAGMVMERWDAESKTATRYVIDRVTARSHTLTLRDEQGEMQTLRLSSLDSR
WSLYQPQTLAVSSGEQLRVLGKMEGLTWRAGDSLQVQQADEAGIQVERQGQMQRLAVGKTPFTAIKVAYG
YVEGLGRSVNDKAQVLAAVAHNDLNQGTLNRLARSGSRIRLYSAQAEDKVAARLGRHPAFSTVTTHLKQV
AGQAGLEEALTRQRVALHTPVQQALHLAVPVLEREGGSLAFSRVSLLAQAQEVAGGQVPLSVLADTIQTQ
IREGALISVDSAPGHGNDLLVSRATFEAEKSILRHIAEGKAAVTPLMANVPESHLAGLTAGQQAATRMIL
ETPDRFTVVQGYAGVGKTTQFRAVMSAIATLPETARPRVVGLGPTHRAVGEMLAAGVESQTLASFLFDTR
QAQQQGEVADFSNTLFLIDESSMLGNSDMASAYALIAAGGGRAVASGDTDQLQAIAPGQPFHLQQTRSAA
DVAIMKEIVRQTPELRPAVYSLIARDVAGALETVERVSPQQVPRQAQAWFPPSSVMEIKKEPDASPDGVD
VVSATLSPTVPATLHDAIVADYLGRTPQARDNTLIITHLNQDRRAVNALIHDARVASGELSGGERTLPIL
VSANIPEGALRRLSTWKTHQDTRPLLDNTYHRIVEVDEKNQLVTLQDETGRQRVLSPQEAVREGVTLYRS
DTITVSVGDRMRFSKSDSERGFVANSTWTISAIEGDSVTLTNGNATRVVHPAIDQAERHIDLAYAITAHG
AQGASESFAIALEGTAGGRRQMVNHASAYVALSRAKQHVQVYTDNREAWVTAIQKADRGATAHEILQPRG
ERTADNAARLLAQAVPLSAGAAGRAVLKQDGLDAGTSLARFVAPGRKYPQPHVAFPAFDENGKAAGAWLS
SLSQGDKPAAVVLPREGRLFGSTAARFVALQASRNGESRLAAGIEEGVKLAARFPDDGIVVRLQGEGVPW
NPGAMTGGRVWADAGETGQGISGEAARLAPEILALRQSEEAARVALEKQAEKTAREMLGQKEKPTPDDVA
PEKMRQIVDEVVKGGVEQERHPADTLPTLPDHQQSAVERVAENLQRERLQQMERELVRDKTLAGD
MMSFSQVKSASNASGYYTDKDNYYVLGSMDDHWYGEGAKQLGLSGAVDKAVFTQLLHGRLPDGADLSRLE
NGVNKHRPGYDLTFSAPKSVSMMAMLGGDRRLIDAHNRAVDVALRQAEALASTRSMRDGVSETVLTGNLI
IARFNHDTSRDQDPQLHTHSVVLNATRNGDKWQALSSDTVGKTGFSENILANRIAIGKIYNQALRSEVEA
MGYQVEVVGKHGLWEMKNVPVEAFSHRSQAIQEAVGADASLKSRDVAALDTRKAKTAIDPTEKMAEWMAT
LKSTGFDIQGYREAAAQRVTQGQVPAPADPAAVDMAKTVRQAISQLSERKVQFTYSDMLAATVSLLPAND
GVIARARAGIDHAIEQQQLIPLDREKGVFTSDIHVLDELSVRTLGQSLGEQGHVTVGAMASSASRHDYSP
LAQQVMTERPPLAMLASPGGAGVQRQRVSELVQATQALGREVQILVSDAKSQRFLQQDAELGQAAIFPQR
VLNDGMAFTPNSTLIVTEGEKLTLKETVTLLDGALRNNVQLLILDTAQRQATGNALSVLRESGIDTLHYA
QTAPVETQILSEPDKPSRFARLADDYVAARQAGDPVVAQVSGPTDQQHLTRAIRSAMKTQGALGQNDTTL
MALEPVWLDSKNRLSRDTYRAGMVMERWDAESKTATRYVIDRVTARSHTLTLRDEQGEMQTLRLSSLDSR
WSLYQPQTLAVSSGEQLRVLGKMEGLTWRAGDSLQVQQADEAGIQVERQGQMQRLAVGKTPFTAIKVAYG
YVEGLGRSVNDKAQVLAAVAHNDLNQGTLNRLARSGSRIRLYSAQAEDKVAARLGRHPAFSTVTTHLKQV
AGQAGLEEALTRQRVALHTPVQQALHLAVPVLEREGGSLAFSRVSLLAQAQEVAGGQVPLSVLADTIQTQ
IREGALISVDSAPGHGNDLLVSRATFEAEKSILRHIAEGKAAVTPLMANVPESHLAGLTAGQQAATRMIL
ETPDRFTVVQGYAGVGKTTQFRAVMSAIATLPETARPRVVGLGPTHRAVGEMLAAGVESQTLASFLFDTR
QAQQQGEVADFSNTLFLIDESSMLGNSDMASAYALIAAGGGRAVASGDTDQLQAIAPGQPFHLQQTRSAA
DVAIMKEIVRQTPELRPAVYSLIARDVAGALETVERVSPQQVPRQAQAWFPPSSVMEIKKEPDASPDGVD
VVSATLSPTVPATLHDAIVADYLGRTPQARDNTLIITHLNQDRRAVNALIHDARVASGELSGGERTLPIL
VSANIPEGALRRLSTWKTHQDTRPLLDNTYHRIVEVDEKNQLVTLQDETGRQRVLSPQEAVREGVTLYRS
DTITVSVGDRMRFSKSDSERGFVANSTWTISAIEGDSVTLTNGNATRVVHPAIDQAERHIDLAYAITAHG
AQGASESFAIALEGTAGGRRQMVNHASAYVALSRAKQHVQVYTDNREAWVTAIQKADRGATAHEILQPRG
ERTADNAARLLAQAVPLSAGAAGRAVLKQDGLDAGTSLARFVAPGRKYPQPHVAFPAFDENGKAAGAWLS
SLSQGDKPAAVVLPREGRLFGSTAARFVALQASRNGESRLAAGIEEGVKLAARFPDDGIVVRLQGEGVPW
NPGAMTGGRVWADAGETGQGISGEAARLAPEILALRQSEEAARVALEKQAEKTAREMLGQKEKPTPDDVA
PEKMRQIVDEVVKGGVEQERHPADTLPTLPDHQQSAVERVAENLQRERLQQMERELVRDKTLAGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6951 | GenBank | WP_035599580 |
| Name | WP_035599580_pET-ATCC-159-1 |
UniProt ID | _ |
| Length | 128 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 128 a.a. Molecular weight: 14737.95 Da Isoelectric Point: 4.8329
>WP_035599580.1 conjugal transfer relaxosome DNA-binding protein TraM [Edwardsiella tarda]
MAKIQAYVSDEVTNKINAIVEARRLEGAKERDVSFSSISTMLLELGLRVYEAQMERKESAFNQMEFNKIL
LENVLKTQFGICKVMGMASLSPYVQGKEKFVYSNMKSDILEDVRQIIEKFFPSEHDDE
MAKIQAYVSDEVTNKINAIVEARRLEGAKERDVSFSSISTMLLELGLRVYEAQMERKESAFNQMEFNKIL
LENVLKTQFGICKVMGMASLSPYVQGKEKFVYSNMKSDILEDVRQIIEKFFPSEHDDE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14264 | GenBank | WP_050979619 |
| Name | traD_DCL27_RS17415_pET-ATCC-159-1 |
UniProt ID | _ |
| Length | 736 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 736 a.a. Molecular weight: 82279.65 Da Isoelectric Point: 5.0782
>WP_050979619.1 type IV conjugative transfer system coupling protein TraD [Edwardsiella tarda]
MSFNAKDMTQGGQIASMRFRMFGQIANIIFYCLFLLFWVIAALTLMARLSWQTFVNGCVYWWCQTLAPMR
SLLRSQPVYHIEYYGRKLAYNAEQILQDKYTIWCGEQLWSSFVLAAIIGGITCLITFFVASWILGHQGKK
QSEDEVTGGRQLTDNPKEVAAMLKRDGRQSDIRIEKLPLIKDSEIQNFCLHGTVGSGKSEFIRRLMNHAR
QRGDMVIVYDRSCEFVKSYYDPAKDKILNPLDVRCANWDLWRECLTQPDFDNVANTLIPMGSSEDPFWQG
SGRTIFAEAAYRMRKDDDRSYAKLVDTLLSIKIDKLRAYLQSSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIECNGESFTIRDWMRSVREDGQNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRQRRV
WIFADELPTLYKLPDLVEILPEARKFGGCYVLGFQSYAQLEDIYGVKGAATLFDVLNTRAFFRSPSKQIA
EFAAGEIGEKEIQKASEQYSYGADPVRDGVSVGKDMERVTLVSYSDIQTLPDLTCFVTLPGPYPAIKLAL
KYQPRPRIAQEFIPRVIDTDAEARLSAVLAAREEEGRSIDGLFESDSQAAAGVQPSQPAVTPVDSQPVNT
VSRKMAATTAAAAVVAAVQGGTDAVAAMGGGVERDLQELQVSAQPEQQDQEDVLPPGIDVQGEVVDPDAY
AAWQAGSHTLRDKQRQEEVNVNHSHVVSDEELGDGF
MSFNAKDMTQGGQIASMRFRMFGQIANIIFYCLFLLFWVIAALTLMARLSWQTFVNGCVYWWCQTLAPMR
SLLRSQPVYHIEYYGRKLAYNAEQILQDKYTIWCGEQLWSSFVLAAIIGGITCLITFFVASWILGHQGKK
QSEDEVTGGRQLTDNPKEVAAMLKRDGRQSDIRIEKLPLIKDSEIQNFCLHGTVGSGKSEFIRRLMNHAR
QRGDMVIVYDRSCEFVKSYYDPAKDKILNPLDVRCANWDLWRECLTQPDFDNVANTLIPMGSSEDPFWQG
SGRTIFAEAAYRMRKDDDRSYAKLVDTLLSIKIDKLRAYLQSSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIECNGESFTIRDWMRSVREDGQNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRQRRV
WIFADELPTLYKLPDLVEILPEARKFGGCYVLGFQSYAQLEDIYGVKGAATLFDVLNTRAFFRSPSKQIA
EFAAGEIGEKEIQKASEQYSYGADPVRDGVSVGKDMERVTLVSYSDIQTLPDLTCFVTLPGPYPAIKLAL
KYQPRPRIAQEFIPRVIDTDAEARLSAVLAAREEEGRSIDGLFESDSQAAAGVQPSQPAVTPVDSQPVNT
VSRKMAATTAAAAVVAAVQGGTDAVAAMGGGVERDLQELQVSAQPEQQDQEDVLPPGIDVQGEVVDPDAY
AAWQAGSHTLRDKQRQEEVNVNHSHVVSDEELGDGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 101661..125471
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DCL27_RS17405 (DCL27_17430) | 100958..101185 | + | 228 | WP_035599335 | type II toxin-antitoxin system VapB family antitoxin | - |
| DCL27_RS17410 (DCL27_17435) | 101185..101583 | + | 399 | WP_035599338 | tRNA(fMet)-specific endonuclease VapC | - |
| DCL27_RS17415 (DCL27_17440) | 101661..103871 | - | 2211 | WP_050979619 | type IV conjugative transfer system coupling protein TraD | virb4 |
| DCL27_RS17420 (DCL27_17445) | 103958..104476 | - | 519 | WP_109691577 | conjugal transfer protein TraS | - |
| DCL27_RS17425 (DCL27_17450) | 104473..107325 | - | 2853 | WP_035599532 | conjugal transfer mating-pair stabilization protein TraG | traG |
| DCL27_RS17430 (DCL27_17455) | 107325..108692 | - | 1368 | WP_035599543 | conjugal transfer pilus assembly protein TraH | traH |
| DCL27_RS17435 (DCL27_17460) | 108689..109228 | - | 540 | WP_035599587 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| DCL27_RS17440 (DCL27_17465) | 109209..109442 | - | 234 | WP_035599544 | type-F conjugative transfer system pilin chaperone TraQ | - |
| DCL27_RS17445 (DCL27_17470) | 109453..110205 | - | 753 | WP_035599547 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| DCL27_RS17450 (DCL27_17475) | 110236..112011 | - | 1776 | WP_050979625 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| DCL27_RS17455 (DCL27_17480) | 112064..112636 | - | 573 | WP_035599590 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| DCL27_RS17460 (DCL27_17485) | 112740..113750 | - | 1011 | WP_050979623 | conjugal transfer pilus assembly protein TraU | traU |
| DCL27_RS17465 (DCL27_17490) | 113747..114382 | - | 636 | WP_035599550 | type-F conjugative transfer system protein TraW | traW |
| DCL27_RS17470 (DCL27_17495) | 114384..114755 | - | 372 | WP_035599552 | type-F conjugative transfer system protein TrbI | - |
| DCL27_RS17475 (DCL27_17500) | 114752..117379 | - | 2628 | WP_035599553 | type IV secretion system protein TraC | virb4 |
| DCL27_RS17480 (DCL27_17505) | 117508..118074 | - | 567 | WP_035599554 | type IV conjugative transfer system lipoprotein TraV | traV |
| DCL27_RS17485 (DCL27_17510) | 118086..118298 | - | 213 | WP_035599555 | hypothetical protein | - |
| DCL27_RS17490 (DCL27_17515) | 118295..118666 | - | 372 | WP_146196432 | hypothetical protein | - |
| DCL27_RS17495 (DCL27_17520) | 118666..120051 | - | 1386 | WP_035599560 | F-type conjugal transfer pilus assembly protein TraB | traB |
| DCL27_RS17500 (DCL27_17525) | 120048..120779 | - | 732 | WP_035599563 | type-F conjugative transfer system secretin TraK | traK |
| DCL27_RS17505 (DCL27_17530) | 120766..121332 | - | 567 | WP_035599565 | type IV conjugative transfer system protein TraE | traE |
| DCL27_RS17510 (DCL27_17535) | 121350..121655 | - | 306 | WP_035599570 | type IV conjugative transfer system protein TraL | traL |
| DCL27_RS17515 (DCL27_17540) | 121671..122018 | - | 348 | WP_035599572 | type IV conjugative transfer system pilin TraA | - |
| DCL27_RS17520 (DCL27_17545) | 122090..122302 | - | 213 | WP_071526360 | TraY domain-containing protein | - |
| DCL27_RS17525 (DCL27_17550) | 122434..123129 | - | 696 | WP_146196433 | hypothetical protein | - |
| DCL27_RS17530 (DCL27_17555) | 123376..124116 | - | 741 | WP_035599577 | complement resistance protein TraT | - |
| DCL27_RS17535 (DCL27_17560) | 124173..124559 | - | 387 | WP_035599580 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| DCL27_RS17540 (DCL27_17565) | 124998..125471 | + | 474 | WP_035599583 | transglycosylase SLT domain-containing protein | virB1 |
| DCL27_RS17545 (DCL27_17570) | 125976..126881 | - | 906 | WP_035599585 | ArdC family protein | - |
| DCL27_RS17550 (DCL27_17575) | 127550..128278 | - | 729 | WP_035601205 | plasmid SOS inhibition protein A | - |
| DCL27_RS17555 (DCL27_17580) | 128275..128706 | - | 432 | WP_035601208 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 19683 | GenBank | NZ_CP084508 |
| Plasmid name | pET-ATCC-159-1 | Incompatibility group | - |
| Plasmid size | 145764 bp | Coordinate of oriT [Strand] | 124871..124906 [+] |
| Host baterium | Edwardsiella tarda ATCC 15947 = NBRC 105688 strain ATCC 15947 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | faeC, faeD, faeE, faeF, faeH, faeI |
| Metal resistance gene | cusR/ylcA, silC, silB, silA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |