Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119228 |
| Name | oriT_pWLK-101716 |
| Organism | Raoultella ornithinolytica strain WLK218 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP038275 (80271..80319 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pWLK-101716
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file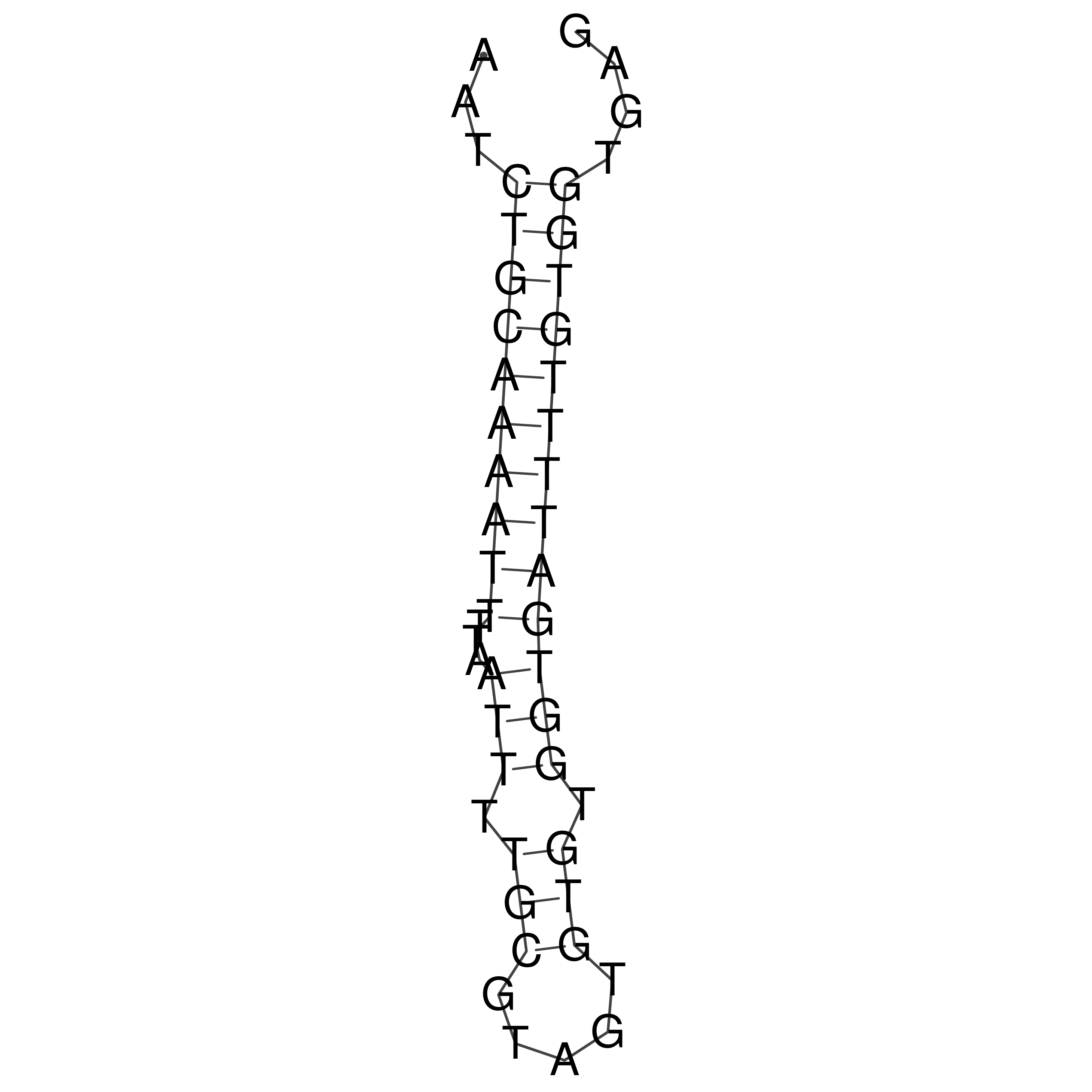
Relaxase
| ID | 12167 | GenBank | WP_137055834 |
| Name | traI_E4K08_RS00300_pWLK-101716 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190598.14 Da Isoelectric Point: 5.5946
>WP_137055834.1 conjugative transfer relaxase/helicase TraI [Raoultella ornithinolytica]
MMSIGSVKSAGSASNYYTDKDNYYVIGSMDERWQGKGAEALGLDGKVDKQVFTDLLKGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSVMAMIGGDKRLIDAHNQAVTEAVKQVETLAATRVMTDGKSETVLTGNLI
VAKFNHDTNRNQEPQLHTHAVVINATQNGGKWQSLGTDTVGKTGFIENVYANQIAFGKLYREVLKPLVEK
LGYETEVTGKHGMWEMKDVPVEPFSTRSQEVREAAGPDASLKSRDVATLDTRKSKETLDPAEKMVEWVNT
LKESGFDIHSYREDADKRAAEINRAPVSPAKTDGPDISDVVTKAIAGLSDRKVQFTYADMLARTVSQLEA
KPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDTAVVRQSPF
SDAVSVLAQDRPTMGIVSGQGGAAGQRERVAELTLMAREQGRDVHIIAADNRSREFLSADARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKMSLKETLSMLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGKQATADIISEPEKGARYSRLAQEFAASVREGQESVAQISGSREQGILNGLIRETLKSEGVLGEKDM
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRQHDRFVIDRVTASSNMLTLRDKEGERLDMKVSAVD
SQWTLFRADKLTVAEGERLAVLGKIPDTRLKGGESVTVLKAEEGQLTVQRPGQKTTQSLSVGSSPFDGIK
VGHGWVESPGRSVSETATVFASLTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIALQKAGLHTPAEQVIHLSIPLLESEELTFTRPQLLATALETGGGKVPMTEIDATIQ
AQIKAGSLLNVPVAPGHGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDSVPGSLMTDLTSGQRASTRM
ILETTDRFTVVQGYAGVGKTTQLRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVNRALATIEQITPEQVPRKDGAWVPENSVVEFTPAQEETIQK
ALNKGESVPAGQPTTLYEALVKDYADRTPEAQSQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKTTRTLNPKADQAQQHVDLAYAITA
HGAQGASEPFAIALEGVAGGRERMASFESSYVGLSRMKQHVQVYTDNREGWIRAINHSPEKATAHDILEP
RNDRAVKTAGQLFSRARALDEAAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDNDGRLQVIGGEGRVMGNEEARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMERQTEQATREVSGEEKKVGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVATESLQRDRLQQMERDMVRDLSREKTLG
GD
MMSIGSVKSAGSASNYYTDKDNYYVIGSMDERWQGKGAEALGLDGKVDKQVFTDLLKGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSVMAMIGGDKRLIDAHNQAVTEAVKQVETLAATRVMTDGKSETVLTGNLI
VAKFNHDTNRNQEPQLHTHAVVINATQNGGKWQSLGTDTVGKTGFIENVYANQIAFGKLYREVLKPLVEK
LGYETEVTGKHGMWEMKDVPVEPFSTRSQEVREAAGPDASLKSRDVATLDTRKSKETLDPAEKMVEWVNT
LKESGFDIHSYREDADKRAAEINRAPVSPAKTDGPDISDVVTKAIAGLSDRKVQFTYADMLARTVSQLEA
KPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDTAVVRQSPF
SDAVSVLAQDRPTMGIVSGQGGAAGQRERVAELTLMAREQGRDVHIIAADNRSREFLSADARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKMSLKETLSMLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGKQATADIISEPEKGARYSRLAQEFAASVREGQESVAQISGSREQGILNGLIRETLKSEGVLGEKDM
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRQHDRFVIDRVTASSNMLTLRDKEGERLDMKVSAVD
SQWTLFRADKLTVAEGERLAVLGKIPDTRLKGGESVTVLKAEEGQLTVQRPGQKTTQSLSVGSSPFDGIK
VGHGWVESPGRSVSETATVFASLTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIALQKAGLHTPAEQVIHLSIPLLESEELTFTRPQLLATALETGGGKVPMTEIDATIQ
AQIKAGSLLNVPVAPGHGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDSVPGSLMTDLTSGQRASTRM
ILETTDRFTVVQGYAGVGKTTQLRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVNRALATIEQITPEQVPRKDGAWVPENSVVEFTPAQEETIQK
ALNKGESVPAGQPTTLYEALVKDYADRTPEAQSQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKTTRTLNPKADQAQQHVDLAYAITA
HGAQGASEPFAIALEGVAGGRERMASFESSYVGLSRMKQHVQVYTDNREGWIRAINHSPEKATAHDILEP
RNDRAVKTAGQLFSRARALDEAAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDNDGRLQVIGGEGRVMGNEEARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMERQTEQATREVSGEEKKVGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVATESLQRDRLQQMERDMVRDLSREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6949 | GenBank | WP_020805752 |
| Name | WP_020805752_pWLK-101716 |
UniProt ID | A0A377TIM3 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14772.81 Da Isoelectric Point: 4.5715
>WP_020805752.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Gammaproteobacteria]
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A377TIM3 |
T4CP
| ID | 14249 | GenBank | WP_032736842 |
| Name | traD_E4K08_RS00305_pWLK-101716 |
UniProt ID | _ |
| Length | 764 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 764 a.a. Molecular weight: 85398.61 Da Isoelectric Point: 5.0675
>WP_032736842.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTISYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTICVVTFFVASWVLGRQGKL
QSEDENTGGRQLSDKPKDVARQMKRDGAASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMRGDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYNPRPKIAEGFIQRRLDTRIDERLSAQLEAREAEGSLVRALFTPDEPVSEPAEEKAGVQPEQTKPAKDS
VLPVPVKTSSMAAEGAGKTPAVPLSGAPVTRVKTVPLIRPKPAASVTGTAAAASTAAVSATAGGTEQELA
QQPVEADQDMLPAGMNEDGVIEDMQAYDAWLEGEQIQRDMQRREEVNINHSRSEQDDIEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTISYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTICVVTFFVASWVLGRQGKL
QSEDENTGGRQLSDKPKDVARQMKRDGAASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMRGDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYNPRPKIAEGFIQRRLDTRIDERLSAQLEAREAEGSLVRALFTPDEPVSEPAEEKAGVQPEQTKPAKDS
VLPVPVKTSSMAAEGAGKTPAVPLSGAPVTRVKTVPLIRPKPAASVTGTAAAASTAAVSATAGGTEQELA
QQPVEADQDMLPAGMNEDGVIEDMQAYDAWLEGEQIQRDMQRREEVNINHSRSEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 54833..77892
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| E4K08_RS00305 (E4K08_00305) | 54833..57127 | - | 2295 | WP_032736842 | type IV conjugative transfer system coupling protein TraD | virb4 |
| E4K08_RS00310 (E4K08_00310) | 57334..57861 | - | 528 | WP_032736843 | hypothetical protein | - |
| E4K08_RS00315 (E4K08_00315) | 57872..60715 | - | 2844 | WP_076752118 | conjugal transfer mating-pair stabilization protein TraG | traG |
| E4K08_RS00320 (E4K08_00320) | 60715..62085 | - | 1371 | WP_032736846 | conjugal transfer pilus assembly protein TraH | traH |
| E4K08_RS00325 (E4K08_00325) | 62072..62635 | - | 564 | WP_224250969 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| E4K08_RS00330 (E4K08_00330) | 62622..62858 | - | 237 | WP_032736848 | type-F conjugative transfer system pilin chaperone TraQ | - |
| E4K08_RS00335 (E4K08_00335) | 62869..63621 | - | 753 | WP_042946285 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| E4K08_RS00340 (E4K08_00340) | 63642..63968 | - | 327 | WP_032736851 | hypothetical protein | - |
| E4K08_RS00345 (E4K08_00345) | 64015..64200 | - | 186 | WP_032736852 | hypothetical protein | - |
| E4K08_RS00350 (E4K08_00350) | 64197..64433 | - | 237 | WP_076752122 | conjugal transfer protein TrbE | - |
| E4K08_RS00355 (E4K08_00355) | 64423..65034 | - | 612 | WP_032736855 | hypothetical protein | - |
| E4K08_RS00360 (E4K08_00360) | 65148..67091 | - | 1944 | WP_032736856 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| E4K08_RS00365 (E4K08_00365) | 67088..67714 | - | 627 | WP_032736857 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| E4K08_RS00370 (E4K08_00370) | 67727..68686 | - | 960 | WP_088912317 | conjugal transfer pilus assembly protein TraU | traU |
| E4K08_RS00375 (E4K08_00375) | 68730..69356 | - | 627 | WP_042946288 | type-F conjugative transfer system protein TraW | traW |
| E4K08_RS00380 (E4K08_00380) | 69353..69742 | - | 390 | WP_032736860 | type-F conjugative transfer system protein TrbI | - |
| E4K08_RS00385 (E4K08_00385) | 69742..72381 | - | 2640 | WP_137055835 | type IV secretion system protein TraC | virb4 |
| E4K08_RS00390 (E4K08_00390) | 72474..72857 | - | 384 | WP_042946290 | hypothetical protein | - |
| E4K08_RS00395 (E4K08_00395) | 72945..73244 | - | 300 | WP_053390308 | hypothetical protein | - |
| E4K08_RS30855 | 73241..73642 | - | 402 | WP_032736864 | hypothetical protein | - |
| E4K08_RS00405 (E4K08_00405) | 73727..74005 | - | 279 | WP_032736865 | hypothetical protein | - |
| E4K08_RS00410 (E4K08_00410) | 74117..74686 | - | 570 | WP_042946293 | type IV conjugative transfer system lipoprotein TraV | traV |
| E4K08_RS30525 | 74706..74867 | - | 162 | WP_154235504 | hypothetical protein | - |
| E4K08_RS00415 (E4K08_00415) | 74860..76280 | - | 1421 | Protein_78 | F-type conjugal transfer pilus assembly protein TraB | - |
| E4K08_RS00420 (E4K08_00420) | 76280..77014 | - | 735 | WP_064343528 | type-F conjugative transfer system secretin TraK | traK |
| E4K08_RS00425 (E4K08_00425) | 77001..77567 | - | 567 | WP_114499520 | type IV conjugative transfer system protein TraE | traE |
| E4K08_RS00430 (E4K08_00430) | 77587..77892 | - | 306 | WP_032736870 | type IV conjugative transfer system protein TraL | traL |
| E4K08_RS00435 (E4K08_00435) | 77906..78274 | - | 369 | WP_064343525 | type IV conjugative transfer system pilin TraA | - |
| E4K08_RS30860 | 78629..79330 | - | 702 | WP_223174962 | hypothetical protein | - |
| E4K08_RS00450 (E4K08_00450) | 79568..79960 | - | 393 | WP_020805752 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| E4K08_RS00455 (E4K08_00455) | 80464..80886 | + | 423 | WP_020315406 | transglycosylase SLT domain-containing protein | - |
| E4K08_RS00460 (E4K08_00460) | 80909..81451 | - | 543 | WP_020314754 | antirestriction protein | - |
| E4K08_RS30865 (E4K08_00465) | 82100..82219 | - | 120 | Protein_87 | SAM-dependent DNA methyltransferase | - |
| E4K08_RS00470 (E4K08_00470) | 82304..82534 | - | 231 | WP_020314756 | hypothetical protein | - |
Host bacterium
| ID | 19658 | GenBank | NZ_CP038275 |
| Plasmid name | pWLK-101716 | Incompatibility group | IncFIA |
| Plasmid size | 101716 bp | Coordinate of oriT [Strand] | 80271..80319 [+] |
| Host baterium | Raoultella ornithinolytica strain WLK218 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |