Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119149 |
| Name | oriT_On28M|unnamed |
| Organism | Citrobacter sp. On28M |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAABKH010000009 (6781..6862 [-], 82 nt) |
| oriT length | 82 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 82 nt
>oriT_On28M|unnamed
GGGACAGGATATGTTTTGTTGAACCGCCTGCACGCAGTCGGCTTCAAAAAACATCCTGGTCAGGGCGAAGCCCCGACACCCC
GGGACAGGATATGTTTTGTTGAACCGCCTGCACGCAGTCGGCTTCAAAAAACATCCTGGTCAGGGCGAAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file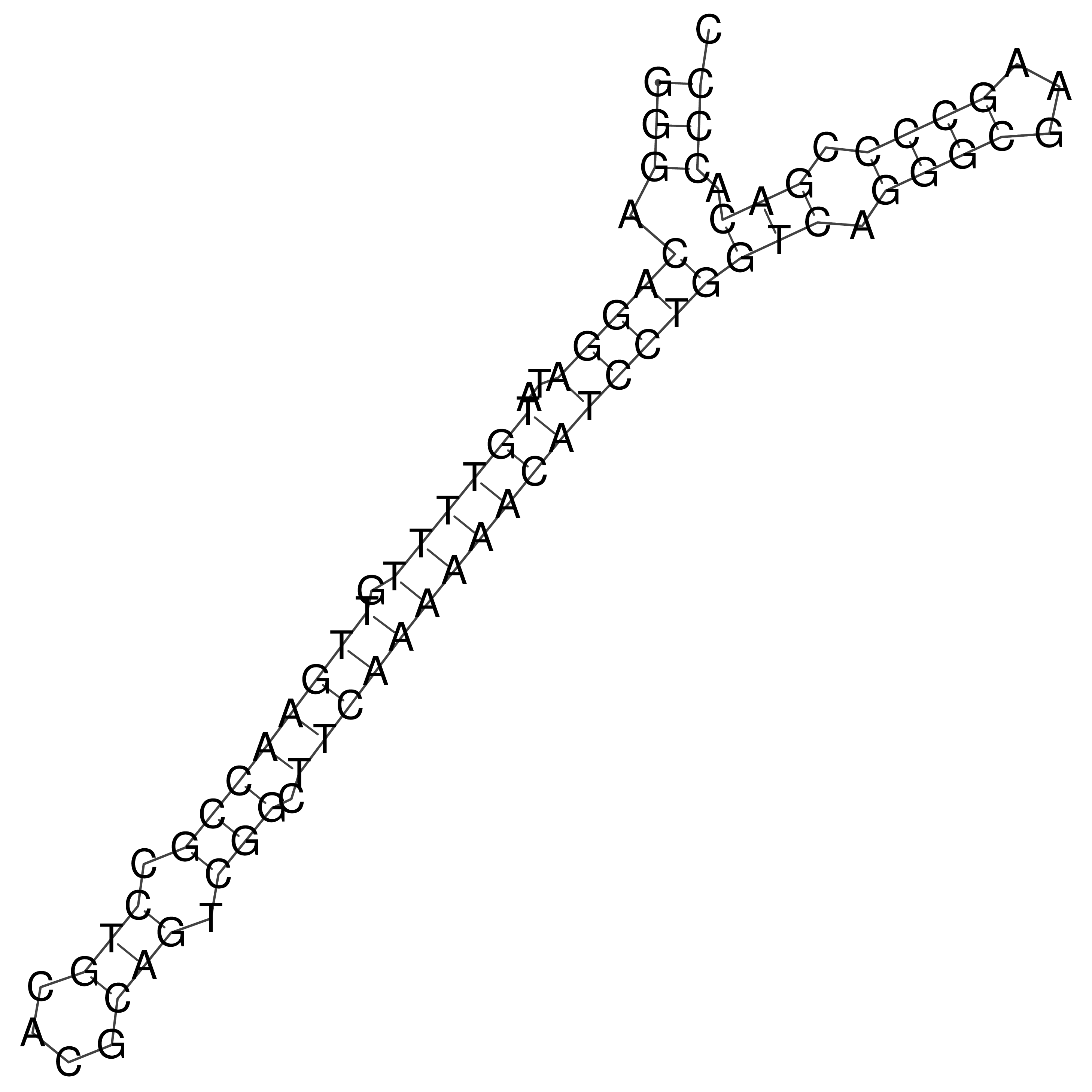
Relaxase
| ID | 12109 | GenBank | WP_219608514 |
| Name | traI_CTBON28M_RS24120_On28M|unnamed |
UniProt ID | _ |
| Length | 663 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 663 a.a. Molecular weight: 76052.27 Da Isoelectric Point: 5.9734
>WP_219608514.1 TraI/MobA(P) family conjugative relaxase [Citrobacter sp. On28M]
MIVIVPEKRRDGKSSFLTLVSYLTLREDVRLAEPVSPDAPVMRMSRSKEAIFDRLVDYIDRSENAVTQTP
VKEFADGRSQVLSGSVACETNCFSLDTASAEMNIVAAQNTRCIDPVYHFILSWPEQDNPVDADIFDCARS
ALHALGMKEHQFVTAIHRDTDHVHCHVAVNRINPVSYRAANLYNDIDTLHKVCRFLEIKYDYTPENGAWV
NENGAVVRNTNDVKSIPRKARQLEYHSDKESLYSYAVNECRERIGEILGSGEASWETLHAELIRAGLDMR
EKGEGLAIYSRHDENVTPIKASTLHPDLTRQCLEKWLGDFEPSPEVVNNFDDDGVYLGSNYPQEFVYDAR
AHVRDQGARSERRLARAVAREDLDARYKAYKNAWVRPKMPDTDSGVQFRALSNRYAWKKEQVRYVFDDPL
IRKLIYRTLELERQKEFEMLRDKVKAEKTQFYSRPENRRLSYWQWVERQAAAQDQAAISRLRGRAYKLKQ
KQRTPGLSENAIVCAVADDIATVNIDGFESRVNRDGTIQYLQDGHVVIQDSGERIEIADPYREEGIHISD
AMLIAEEKSGERLLFSGEGRFVNSACDVVQWFNDGGEKPLPLTDPQQRVMAGYDKPETHSDRPVMTHRIV
DEETYLASRQGARAVDDRETEELKPEKKTHRLR
MIVIVPEKRRDGKSSFLTLVSYLTLREDVRLAEPVSPDAPVMRMSRSKEAIFDRLVDYIDRSENAVTQTP
VKEFADGRSQVLSGSVACETNCFSLDTASAEMNIVAAQNTRCIDPVYHFILSWPEQDNPVDADIFDCARS
ALHALGMKEHQFVTAIHRDTDHVHCHVAVNRINPVSYRAANLYNDIDTLHKVCRFLEIKYDYTPENGAWV
NENGAVVRNTNDVKSIPRKARQLEYHSDKESLYSYAVNECRERIGEILGSGEASWETLHAELIRAGLDMR
EKGEGLAIYSRHDENVTPIKASTLHPDLTRQCLEKWLGDFEPSPEVVNNFDDDGVYLGSNYPQEFVYDAR
AHVRDQGARSERRLARAVAREDLDARYKAYKNAWVRPKMPDTDSGVQFRALSNRYAWKKEQVRYVFDDPL
IRKLIYRTLELERQKEFEMLRDKVKAEKTQFYSRPENRRLSYWQWVERQAAAQDQAAISRLRGRAYKLKQ
KQRTPGLSENAIVCAVADDIATVNIDGFESRVNRDGTIQYLQDGHVVIQDSGERIEIADPYREEGIHISD
AMLIAEEKSGERLLFSGEGRFVNSACDVVQWFNDGGEKPLPLTDPQQRVMAGYDKPETHSDRPVMTHRIV
DEETYLASRQGARAVDDRETEELKPEKKTHRLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14184 | GenBank | WP_219608589 |
| Name | trbC_CTBON28M_RS24675_On28M|unnamed |
UniProt ID | _ |
| Length | 731 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 731 a.a. Molecular weight: 83078.44 Da Isoelectric Point: 8.0317
>WP_219608589.1 F-type conjugative transfer protein TrbC [Citrobacter sp. On28M]
MTTGNTQPVDKQRVRQITQSTWIDDYLLAPATVQAGLVVILVLTFLRTWILLPGVFATSIWLLTFFGQLF
RMPLRMPKDIGGFDMTTERENALEFKGPMGLFRFTRRRLTWEKSAGIMCLGHARRRFLGRELWLTRDDCL
RHMQLLATTGSGKTEALLSLQLNSLCMGRGMMFSDGKAETKLAYSIWSLMRRYGQEDNYYVLNFLNGGRD
RFDELLGNDRTRPQSNSINFFGDATATFIIQLMESLLPQVGSNEAGWQDKAKPMLYGLVYALYYKCRKEN
IRLSQSMIQKHLPLKEMADLYIEGKQNGWHDEAVSALEAYLSNLAGFRMELVSKSSEWDQGVYDQHGFLT
QQFSRMLAMFNDVYGHIFSSDAGDIDISDVLHNDRTLVVLIPALELSKSESANLGKLYISAKRMVYARDL
GYQLEGKSRDVLTSAKFYNPFPFTDAHDELASYFAPGMDDLAAQMRSLGVMLVISAQDIQRFVAQFKGEY
QTVNANTLVKWFMAMQDEKDTFELAKATAGKDYYAELGEMKRTPGTVSSSYEEANTTYIREKNRITLDEL
KDLNPGEGFISFKSALVPSSAIYIPDSEKISSKLSLRINRFIDTGKPTEADLVAQNPHLRRLLPPSELEV
SMLLQRTDEIIAGNETHTLPPLMDPILARLMKVALDLDNRCDISYTPTQRGILLFEAARDVLKKRGKKWF
TVKTQPNQLRVSDAMAHRLARKSTAFMPQEQ
MTTGNTQPVDKQRVRQITQSTWIDDYLLAPATVQAGLVVILVLTFLRTWILLPGVFATSIWLLTFFGQLF
RMPLRMPKDIGGFDMTTERENALEFKGPMGLFRFTRRRLTWEKSAGIMCLGHARRRFLGRELWLTRDDCL
RHMQLLATTGSGKTEALLSLQLNSLCMGRGMMFSDGKAETKLAYSIWSLMRRYGQEDNYYVLNFLNGGRD
RFDELLGNDRTRPQSNSINFFGDATATFIIQLMESLLPQVGSNEAGWQDKAKPMLYGLVYALYYKCRKEN
IRLSQSMIQKHLPLKEMADLYIEGKQNGWHDEAVSALEAYLSNLAGFRMELVSKSSEWDQGVYDQHGFLT
QQFSRMLAMFNDVYGHIFSSDAGDIDISDVLHNDRTLVVLIPALELSKSESANLGKLYISAKRMVYARDL
GYQLEGKSRDVLTSAKFYNPFPFTDAHDELASYFAPGMDDLAAQMRSLGVMLVISAQDIQRFVAQFKGEY
QTVNANTLVKWFMAMQDEKDTFELAKATAGKDYYAELGEMKRTPGTVSSSYEEANTTYIREKNRITLDEL
KDLNPGEGFISFKSALVPSSAIYIPDSEKISSKLSLRINRFIDTGKPTEADLVAQNPHLRRLLPPSELEV
SMLLQRTDEIIAGNETHTLPPLMDPILARLMKVALDLDNRCDISYTPTQRGILLFEAARDVLKKRGKKWF
TVKTQPNQLRVSDAMAHRLARKSTAFMPQEQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 19579 | GenBank | NZ_JAABKH010000009 |
| Plasmid name | On28M|unnamed | Incompatibility group | IncFIB |
| Plasmid size | 107461 bp | Coordinate of oriT [Strand] | 6781..6862 [-] |
| Host baterium | Citrobacter sp. On28M |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | paa |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIB |