Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119113 |
| Name | oriT_pSM51_Rh08 |
| Organism | Rhizobium beringeri strain SM51 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SILG01000005 (181238..181275 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pSM51_Rh08
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file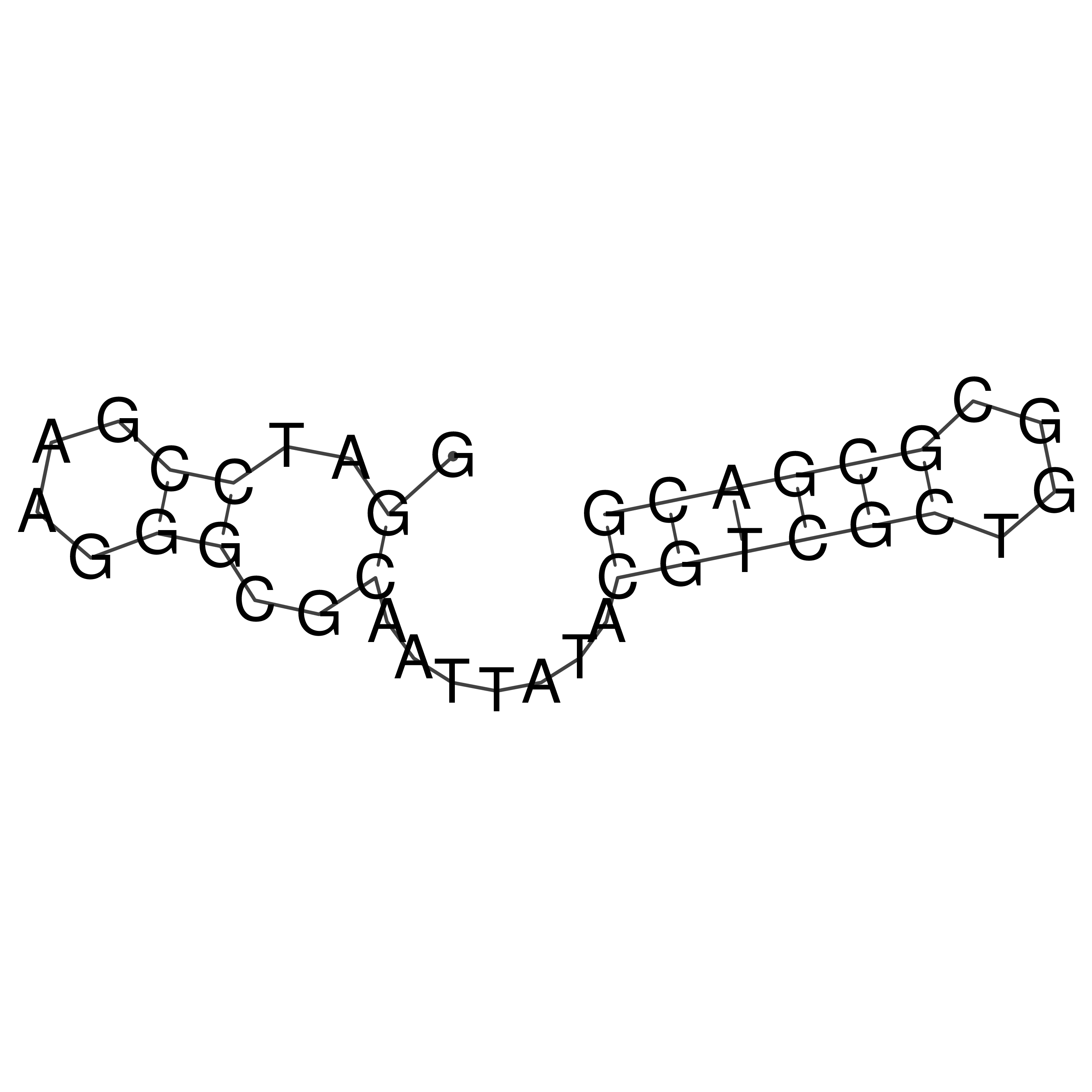
Relaxase
| ID | 12072 | GenBank | WP_130806924 |
| Name | traA_ELH03_RS33635_pSM51_Rh08 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122919.18 Da Isoelectric Point: 9.8379
>WP_130806924.1 Ti-type conjugative transfer relaxase TraA [Rhizobium beringeri]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLIEDGFGSKKVAILGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPTKYTTREM
IRIEAEMANHAIWLSARASHGVREAVLHATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNRLDDKTVIVLDEAGMVSSRQM
ALLVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLARVLEAASGRIVAEIGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYQ
KSALYRQALRFAEARGLQLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIRDTI
KEAKPMVSGITTFPKSLDQAVEGKLAADPSLKKQWEDVSTRFHLVYAQPEAAFKAINVDAMLTDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLIEDGFGSKKVAILGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPTKYTTREM
IRIEAEMANHAIWLSARASHGVREAVLHATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNRLDDKTVIVLDEAGMVSSRQM
ALLVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLARVLEAASGRIVAEIGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYQ
KSALYRQALRFAEARGLQLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIRDTI
KEAKPMVSGITTFPKSLDQAVEGKLAADPSLKKQWEDVSTRFHLVYAQPEAAFKAINVDAMLTDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14136 | GenBank | WP_130806922 |
| Name | traG_ELH03_RS33620_pSM51_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69446.66 Da Isoelectric Point: 9.3616
>WP_130806922.1 Ti-type conjugative transfer system protein TraG [Rhizobium beringeri]
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYGASAAIGILFLFASAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTKALNADKSVLAYLDPATAIGAGVALMCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLLPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGASG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
QEQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIRPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYGASAAIGILFLFASAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTKALNADKSVLAYLDPATAIGAGVALMCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLLPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGASG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
QEQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIRPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14137 | GenBank | WP_130806925 |
| Name | t4cp2_ELH03_RS33720_pSM51_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91431.16 Da Isoelectric Point: 5.7018
>WP_130806925.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLESTRKKWQQKVRPFFEQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAAPLLK
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLESTRKKWQQKVRPFFEQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAAPLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 186411..200547
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH03_RS33640 (ELH03_33640) | 184678..185241 | + | 564 | WP_130714114 | conjugative transfer signal peptidase TraF | - |
| ELH03_RS33645 (ELH03_33645) | 185231..186394 | + | 1164 | WP_130729903 | conjugal transfer protein TraB | - |
| ELH03_RS33650 (ELH03_33650) | 186411..187028 | + | 618 | WP_130654292 | TraH family protein | virB1 |
| ELH03_RS33655 (ELH03_33655) | 187131..187997 | - | 867 | Protein_174 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELH03_RS33660 (ELH03_33660) | 187990..188607 | - | 618 | WP_130654291 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELH03_RS38280 (ELH03_33665) | 188698..189129 | - | 432 | Protein_176 | hypothetical protein | - |
| ELH03_RS33670 (ELH03_33670) | 189232..189474 | - | 243 | WP_130654290 | helix-turn-helix domain-containing protein | - |
| ELH03_RS33675 (ELH03_33675) | 189697..190020 | + | 324 | WP_130654289 | transcriptional repressor TraM | - |
| ELH03_RS33680 (ELH03_33680) | 190024..190728 | - | 705 | WP_130654288 | autoinducer binding domain-containing protein | - |
| ELH03_RS33685 (ELH03_33685) | 191040..192338 | - | 1299 | WP_130654287 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH03_RS33690 (ELH03_33690) | 192350..192790 | - | 441 | WP_130654286 | conjugal transfer protein TrbH | - |
| ELH03_RS33695 (ELH03_33695) | 192794..193606 | - | 813 | WP_130654285 | P-type conjugative transfer protein TrbG | virB9 |
| ELH03_RS33700 (ELH03_33700) | 193624..194286 | - | 663 | WP_130654284 | conjugal transfer protein TrbF | virB8 |
| ELH03_RS33705 (ELH03_33705) | 194307..195494 | - | 1188 | WP_130654283 | P-type conjugative transfer protein TrbL | virB6 |
| ELH03_RS33710 (ELH03_33710) | 195499..195675 | - | 177 | WP_130654282 | entry exclusion protein TrbK | - |
| ELH03_RS33715 (ELH03_33715) | 195669..196466 | - | 798 | WP_130654281 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH03_RS33720 (ELH03_33720) | 196444..198894 | - | 2451 | WP_130806925 | conjugal transfer protein TrbE | virb4 |
| ELH03_RS33725 (ELH03_33725) | 198905..199204 | - | 300 | WP_130654279 | conjugal transfer protein TrbD | virB3 |
| ELH03_RS33730 (ELH03_33730) | 199197..199580 | - | 384 | WP_065276633 | TrbC/VirB2 family protein | virB2 |
| ELH03_RS33735 (ELH03_33735) | 199570..200547 | - | 978 | WP_130654278 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH03_RS33740 (ELH03_33740) | 200544..201182 | - | 639 | WP_130654277 | acyl-homoserine-lactone synthase | - |
| ELH03_RS33745 (ELH03_33745) | 201559..202782 | + | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELH03_RS33750 (ELH03_33750) | 202779..203816 | + | 1038 | WP_130657451 | plasmid partitioning protein RepB | - |
| ELH03_RS33755 (ELH03_33755) | 204006..205220 | + | 1215 | WP_130806927 | plasmid replication protein RepC | - |
Host bacterium
| ID | 19545 | GenBank | NZ_SILG01000005 |
| Plasmid name | pSM51_Rh08 | Incompatibility group | - |
| Plasmid size | 283601 bp | Coordinate of oriT [Strand] | 181238..181275 [+] |
| Host baterium | Rhizobium beringeri strain SM51 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodM, nodE, nodF, nodD, nodA, nodC, nodI, nodJ, nifT, nifA, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE, nifN, fixN, fixO, fixQ, fixG |
| Anti-CRISPR | - |