Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119101 |
| Name | oriT_p201603898-2 |
| Organism | Shigella sonnei strain 201603898 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OP038269 (45127..45199 [-], 73 nt) |
| oriT length | 73 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 73 nt
>oriT_p201603898-2
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file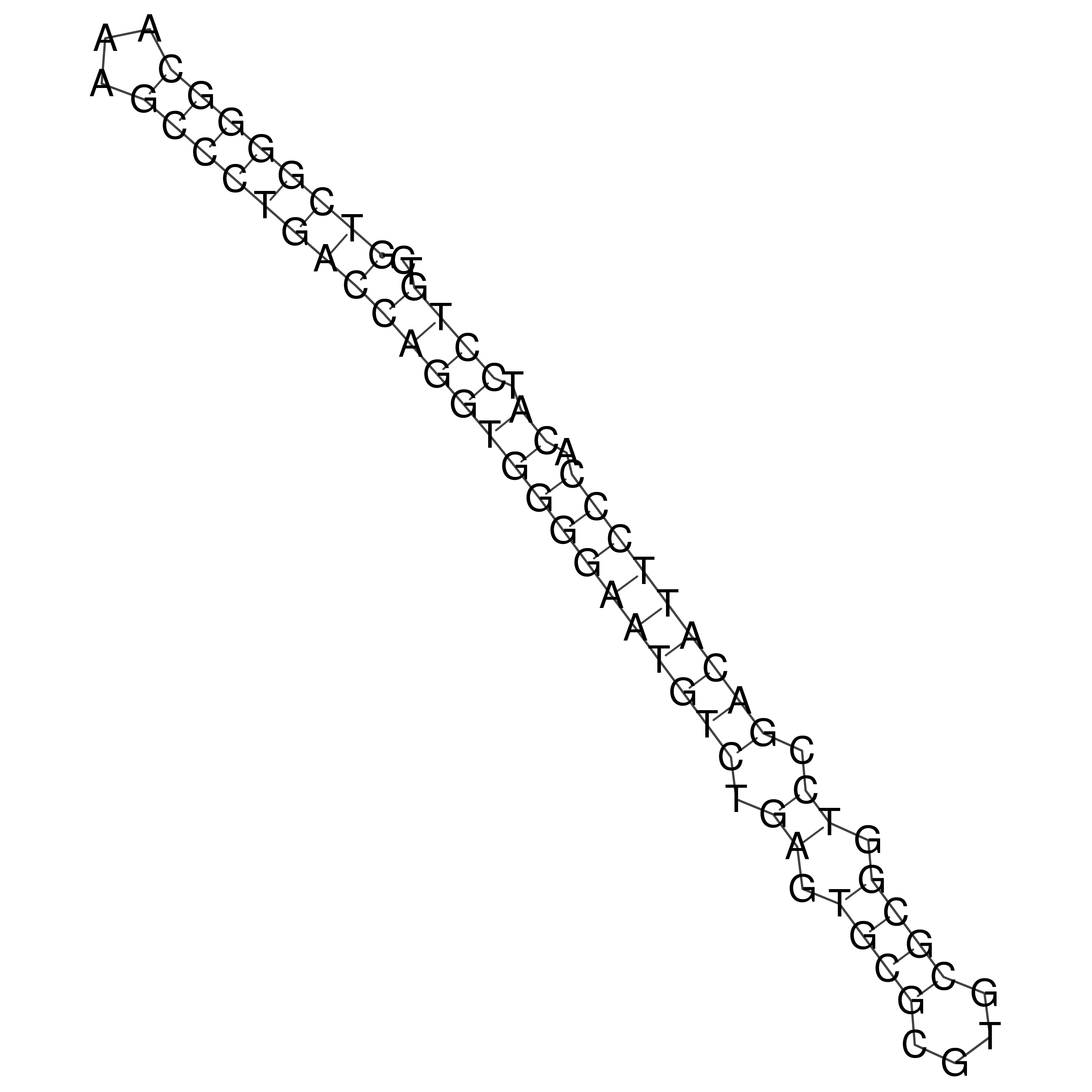
Relaxase
| ID | 12063 | GenBank | WP_052984280 |
| Name | Relaxase_OVP32_RS00295_p201603898-2 |
UniProt ID | _ |
| Length | 903 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 903 a.a. Molecular weight: 104663.00 Da Isoelectric Point: 7.4180
>WP_052984280.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRYFAERLPEMARNGELESCLDVHRTLAKAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGRNLGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
TTVQCESLIEDMIAGGNFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEL
QAGPFEIAGADIFERVKPECRYNPELAASDEVEPGFRRDPVLRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAVQGDRAALSQLRGWDYRDRRKDKRRTTNADRCVILCEPGGTPLYEDTGVLEARLQKDGS
VRFRDRRNGELVCVDYGDRVVFYHHQDRNELVDKLNLIAPVLFDREPGMGFEPEGSYQQFNDVFAEMVAW
HNAAGITGNGHFVISRPDVDLHRQRSEQYYHEYIRQQKSISGGNDTSYTPVQDNEWTPPSPGM
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRYFAERLPEMARNGELESCLDVHRTLAKAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGRNLGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
TTVQCESLIEDMIAGGNFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEL
QAGPFEIAGADIFERVKPECRYNPELAASDEVEPGFRRDPVLRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAVQGDRAALSQLRGWDYRDRRKDKRRTTNADRCVILCEPGGTPLYEDTGVLEARLQKDGS
VRFRDRRNGELVCVDYGDRVVFYHHQDRNELVDKLNLIAPVLFDREPGMGFEPEGSYQQFNDVFAEMVAW
HNAAGITGNGHFVISRPDVDLHRQRSEQYYHEYIRQQKSISGGNDTSYTPVQDNEWTPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6897 | GenBank | WP_001291058 |
| Name | WP_001291058_p201603898-2 |
UniProt ID | A0A7I0L241 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12730.52 Da Isoelectric Point: 9.9656
>WP_001291058.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0L241 |
T4CP
| ID | 14126 | GenBank | WP_000077663 |
| Name | trbC_OVP32_RS00320_p201603898-2 |
UniProt ID | _ |
| Length | 767 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 767 a.a. Molecular weight: 87012.13 Da Isoelectric Point: 7.3008
>WP_000077663.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHVM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEMLLPVPDLTASPPHKKNVAQTPSGGKQESRKKRF
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHVM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEMLLPVPDLTASPPHKKNVAQTPSGGKQESRKKRF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 53053..91858
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OVP32_RS00300 (p201603898-2__00061) | 48343..48705 | + | 363 | Protein_61 | cell envelope integrity TolA C-terminal domain-containing protein | - |
| OVP32_RS00305 (p201603898-2__00062) | 48773..49753 | + | 981 | WP_052990530 | IS5-like element ISKpn26 family transposase | - |
| OVP32_RS00310 | 49903..49974 | + | 72 | Protein_63 | tolA family protein | - |
| OVP32_RS00315 | 50072..50302 | - | 231 | WP_065406859 | hypothetical protein | - |
| OVP32_RS00320 (p201603898-2__00065) | 50769..53072 | - | 2304 | WP_000077663 | F-type conjugative transfer protein TrbC | - |
| OVP32_RS00325 (p201603898-2__00066) | 53053..54177 | - | 1125 | WP_000152381 | DsbC family protein | trbB |
| OVP32_RS00330 (p201603898-2__00067) | 54174..55454 | - | 1281 | WP_052973421 | IncI1-type conjugal transfer protein TrbA | trbA |
| OVP32_RS00595 | 56251..56346 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| OVP32_RS00340 (p201603898-2__00069) | 56409..56585 | - | 177 | WP_001054907 | hypothetical protein | - |
| OVP32_RS00345 (p201603898-2__00070) | 56735..57202 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| OVP32_RS00350 (p201603898-2__00071) | 57361..57984 | + | 624 | WP_001155247 | helix-turn-helix domain-containing protein | - |
| OVP32_RS00355 (p201603898-2__00073) | 58182..58397 | - | 216 | WP_000266543 | hypothetical protein | - |
| OVP32_RS00360 (p201603898-2__00074) | 58401..58769 | - | 369 | WP_001345852 | hypothetical protein | - |
| OVP32_RS00365 (p201603898-2__00075) | 59060..59302 | - | 243 | WP_000650923 | hypothetical protein | - |
| OVP32_RS00370 (p201603898-2__00076) | 59632..59784 | + | 153 | WP_001335079 | Hok/Gef family protein | - |
| OVP32_RS00375 (p201603898-2__00077) | 59922..61508 | - | 1587 | WP_000004564 | TnsA endonuclease N-terminal domain-containing protein | - |
| OVP32_RS00380 (p201603898-2__00079) | 61782..62435 | - | 654 | WP_001401768 | plasmid IncI1-type surface exclusion protein ExcA | - |
| OVP32_RS00385 (p201603898-2__00080) | 62523..64688 | - | 2166 | WP_000691808 | DotA/TraY family protein | traY |
| OVP32_RS00390 (p201603898-2__00081) | 64763..65332 | - | 570 | WP_000650752 | conjugal transfer protein TraX | - |
| OVP32_RS00395 (p201603898-2__00082) | 65329..66534 | - | 1206 | WP_001189548 | conjugal transfer protein TraW | traW |
| OVP32_RS00400 (p201603898-2__00083) | 66492..67112 | - | 621 | WP_000286854 | hypothetical protein | traV |
| OVP32_RS00405 (p201603898-2__00084) | 67112..70156 | - | 3045 | WP_001024766 | conjugal transfer protein | traU |
| OVP32_RS00410 (p201603898-2__00085) | 70451..71269 | - | 819 | WP_000753550 | hypothetical protein | traT |
| OVP32_RS00600 (p201603898-2__00086) | 71184..71468 | - | 285 | WP_001353683 | hypothetical protein | - |
| OVP32_RS00420 (p201603898-2__00087) | 71492..71890 | - | 399 | WP_000088813 | DUF6750 family protein | traR |
| OVP32_RS00425 (p201603898-2__00088) | 71937..72467 | - | 531 | WP_000987023 | conjugal transfer protein TraQ | traQ |
| OVP32_RS00430 (p201603898-2__00089) | 72464..73177 | - | 714 | WP_000787003 | conjugal transfer protein TraP | traP |
| OVP32_RS00435 (p201603898-2__00090) | 73174..74505 | - | 1332 | WP_001288254 | conjugal transfer protein TraO | traO |
| OVP32_RS00440 (p201603898-2__00091) | 74509..75483 | - | 975 | WP_000547383 | DotH/IcmK family type IV secretion protein | traN |
| OVP32_RS00445 (p201603898-2__00092) | 75494..76189 | - | 696 | WP_001287545 | DotI/IcmL family type IV secretion protein | traM |
| OVP32_RS00450 (p201603898-2__00093) | 76201..76551 | - | 351 | WP_001056367 | hypothetical protein | traL |
| OVP32_RS00455 (p201603898-2__00094) | 76568..80629 | - | 4062 | WP_000842447 | LPD7 domain-containing protein | - |
| OVP32_RS00460 (p201603898-2__00095) | 80693..80983 | - | 291 | WP_000817801 | IcmT/TraK family protein | traK |
| OVP32_RS00465 (p201603898-2__00096) | 80980..82128 | - | 1149 | WP_000775210 | plasmid transfer ATPase TraJ | virB11 |
| OVP32_RS00470 (p201603898-2__00097) | 82112..82948 | - | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| OVP32_RS00475 (p201603898-2__00098) | 82945..83403 | - | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| OVP32_RS00480 (p201603898-2__00099) | 83507..84709 | - | 1203 | WP_000979993 | conjugal transfer protein TraF | - |
| OVP32_RS00485 (p201603898-2__00100) | 84811..85632 | - | 822 | WP_000845373 | hypothetical protein | traE |
| OVP32_RS00490 | 85797..85991 | - | 195 | Protein_99 | integrase | - |
| OVP32_RS00495 (p201603898-2__00101) | 86105..87466 | - | 1362 | WP_001168171 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| OVP32_RS00500 (p201603898-2__00102) | 87484..88020 | - | 537 | WP_023649457 | prepilin peptidase | - |
| OVP32_RS00505 (p201603898-2__00103) | 88126..88611 | - | 486 | WP_063612151 | lytic transglycosylase domain-containing protein | virB1 |
| OVP32_RS00510 (p201603898-2__00104) | 88656..89192 | - | 537 | WP_000111534 | type 4 pilus major pilin | - |
| OVP32_RS00515 (p201603898-2__00105) | 89254..90348 | - | 1095 | WP_000697158 | type II secretion system F family protein | - |
| OVP32_RS00520 (p201603898-2__00106) | 90350..91858 | - | 1509 | WP_000880807 | GspE/PulE family protein | virB11 |
| OVP32_RS00525 (p201603898-2__00107) | 91961..92419 | - | 459 | WP_000095885 | type IV pilus biogenesis protein PilP | - |
| OVP32_RS00530 (p201603898-2__00108) | 92409..93704 | - | 1296 | WP_000129883 | type 4b pilus protein PilO2 | - |
| OVP32_RS00535 (p201603898-2__00109) | 93725..95344 | - | 1620 | WP_001035562 | PilN family type IVB pilus formation outer membrane protein | - |
| OVP32_RS00540 (p201603898-2__00110) | 95376..95813 | - | 438 | WP_000539952 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 19533 | GenBank | NZ_OP038269 |
| Plasmid name | p201603898-2 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 102642 bp | Coordinate of oriT [Strand] | 45127..45199 [-] |
| Host baterium | Shigella sonnei strain 201603898 |
Cargo genes
| Drug resistance gene | mph(A), blaCTX-M-55, aac(3)-IIa |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |