Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119099 |
| Name | oriT_pSM51_Rh01 |
| Organism | Rhizobium beringeri strain SM51 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SILG01000002 (802876..802904 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSM51_Rh01
AGGGCGCAATATACGTCGCTATCGCGACG
AGGGCGCAATATACGTCGCTATCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file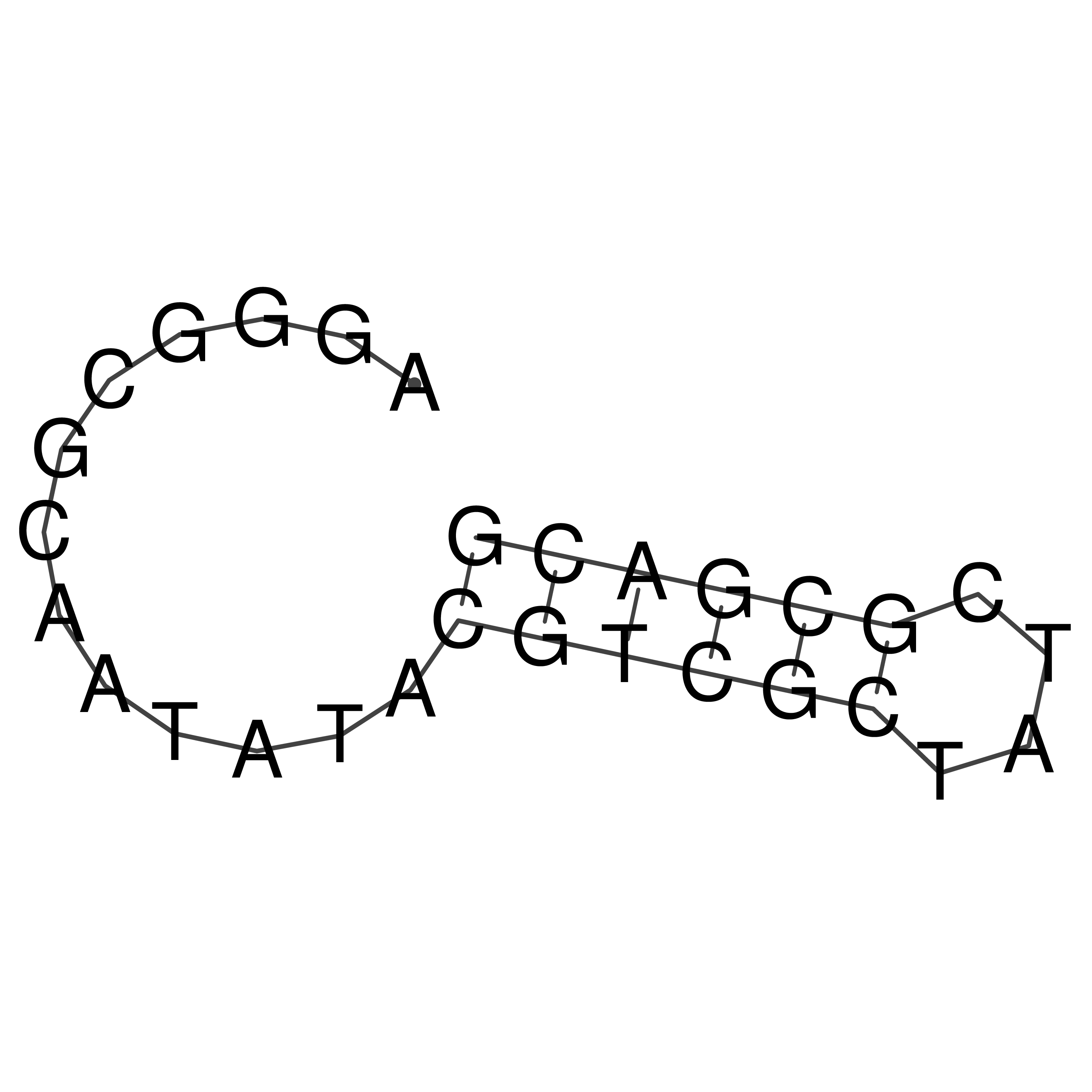
Relaxase
| ID | 12061 | GenBank | WP_130806825 |
| Name | traA_ELH03_RS27340_pSM51_Rh01 |
UniProt ID | _ |
| Length | 1535 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1535 a.a. Molecular weight: 169596.49 Da Isoelectric Point: 7.5061
>WP_130806825.1 Ti-type conjugative transfer relaxase TraA [Rhizobium beringeri]
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQMPTWLRTAIDG
RSVAGASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRSLTEEGFGPKKVAVAGEDGKPLRVVTPDRPKGRIVYKLWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRPYADQGLDGIAQRHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPALLLKQ
LSNERSTFDERDIAKALHRTVDDPAVFANIRARLMASDDLIMLKPQQLDAETGKASEPAVFTTREVLRIE
YDMARSAQLLAECRGFGVSDRVVAAAIRHVETRDTEKSFRLDPEQVDAVRHVTGDNGITAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELAWANGRELPERGNVLVIDEAGMVS
SQQLARVLDIAEQAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRHQWAREASRLFARGE
VEQGLDAYARHGHLIEAGTRDEVIGRIVGDWTDARRQAIRNSASAGNDGRLRGDELLVLAHTNEDVKRLN
EALRSVMSQEGALSENRAFRTERGAREFAVGDRIIFLENARFLEPRAPRLGPQHVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTLVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFEAKPEWGRKPRVDHAAGVTGALVESGQAKFRPNDEDADDSPYADVKTDDGTIHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVERVTIKVAIVDEQTGQKRFEEREVDRNVWTAKRIETSEARQQRIERESHRP
DLFKQLVERLSRSGAKTTTLDFESEAGYRAQAQDFARRRGLDHLSLAAAAVEESLSRRWAGIAERREQLA
KLWERAGVAFGFTIERERRVAYNEERAEPQPVEADVVGSRYLIPPATGFARSADEDALLAQLASPAWKER
EAILRPLLAKIYRDPDAALVALNALSSDTSIEPRRLADDLANAPDRLGRLRGSDLIIDGRAARDERNAAI
TALTDLLPLARAHATEFRRNGERFDLGEQRRRTHMSLPIPALSEKAMARLTEIEAVRRQGGDDAYKTAFA
LATEDRSIVQEVKAVSEALSARFGWSAFTAKADAVAERNMIERMPEDLTSISGEKLTRLFEAVKRFADEQ
HLVERWDRSKIVAAASVDPQKENVPMLAAVTEFKTPVDEEARSRALATPLYRHQRAALAAVASTIWRDPA
GAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLVDRMLAAGRERKEVVQAVPEAGARLRALGSA
YVNLLDAERQATTEERRRMAVAIPGLSTAAEDVLMRLTAEAKKNGRKLSGSAASLGPDIRREFAAVSSAL
DERFGRSAIIRGEKDLSNRVPPAQHRAFEVMQEKLKVLQQTVRRESSEQIISERRQRTTNRSIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQMPTWLRTAIDG
RSVAGASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRSLTEEGFGPKKVAVAGEDGKPLRVVTPDRPKGRIVYKLWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRPYADQGLDGIAQRHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPALLLKQ
LSNERSTFDERDIAKALHRTVDDPAVFANIRARLMASDDLIMLKPQQLDAETGKASEPAVFTTREVLRIE
YDMARSAQLLAECRGFGVSDRVVAAAIRHVETRDTEKSFRLDPEQVDAVRHVTGDNGITAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELAWANGRELPERGNVLVIDEAGMVS
SQQLARVLDIAEQAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRHQWAREASRLFARGE
VEQGLDAYARHGHLIEAGTRDEVIGRIVGDWTDARRQAIRNSASAGNDGRLRGDELLVLAHTNEDVKRLN
EALRSVMSQEGALSENRAFRTERGAREFAVGDRIIFLENARFLEPRAPRLGPQHVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTLVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFEAKPEWGRKPRVDHAAGVTGALVESGQAKFRPNDEDADDSPYADVKTDDGTIHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVERVTIKVAIVDEQTGQKRFEEREVDRNVWTAKRIETSEARQQRIERESHRP
DLFKQLVERLSRSGAKTTTLDFESEAGYRAQAQDFARRRGLDHLSLAAAAVEESLSRRWAGIAERREQLA
KLWERAGVAFGFTIERERRVAYNEERAEPQPVEADVVGSRYLIPPATGFARSADEDALLAQLASPAWKER
EAILRPLLAKIYRDPDAALVALNALSSDTSIEPRRLADDLANAPDRLGRLRGSDLIIDGRAARDERNAAI
TALTDLLPLARAHATEFRRNGERFDLGEQRRRTHMSLPIPALSEKAMARLTEIEAVRRQGGDDAYKTAFA
LATEDRSIVQEVKAVSEALSARFGWSAFTAKADAVAERNMIERMPEDLTSISGEKLTRLFEAVKRFADEQ
HLVERWDRSKIVAAASVDPQKENVPMLAAVTEFKTPVDEEARSRALATPLYRHQRAALAAVASTIWRDPA
GAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLVDRMLAAGRERKEVVQAVPEAGARLRALGSA
YVNLLDAERQATTEERRRMAVAIPGLSTAAEDVLMRLTAEAKKNGRKLSGSAASLGPDIRREFAAVSSAL
DERFGRSAIIRGEKDLSNRVPPAQHRAFEVMQEKLKVLQQTVRRESSEQIISERRQRTTNRSIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8692..18252
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH03_RS23675 (ELH03_23675) | 5201..5617 | - | 417 | WP_130660238 | UPF0158 family protein | - |
| ELH03_RS23680 (ELH03_23680) | 5961..6869 | + | 909 | WP_130766952 | nucleotidyltransferase and HEPN domain-containing protein | - |
| ELH03_RS38015 (ELH03_23685) | 7089..7175 | - | 87 | Protein_6 | SDR family oxidoreductase | - |
| ELH03_RS23690 (ELH03_23690) | 7182..7514 | + | 333 | Protein_7 | LysR substrate-binding domain-containing protein | - |
| ELH03_RS23695 (ELH03_23695) | 7762..7977 | + | 216 | WP_130766736 | DUF3072 domain-containing protein | - |
| ELH03_RS23705 (ELH03_23705) | 8692..9687 | - | 996 | WP_130766737 | P-type DNA transfer ATPase VirB11 | virB11 |
| ELH03_RS23710 (ELH03_23710) | 9696..10868 | - | 1173 | WP_130766738 | type IV secretion system protein VirB10 | virB10 |
| ELH03_RS23715 (ELH03_23715) | 10878..11732 | - | 855 | WP_130766739 | P-type conjugative transfer protein VirB9 | virB9 |
| ELH03_RS23720 (ELH03_23720) | 11729..12400 | - | 672 | WP_130766740 | virB8 family protein | virB8 |
| ELH03_RS23725 (ELH03_23725) | 12402..12686 | - | 285 | WP_130666527 | hypothetical protein | - |
| ELH03_RS23730 (ELH03_23730) | 12739..13659 | - | 921 | WP_130766741 | type IV secretion system protein | virB6 |
| ELH03_RS23735 (ELH03_23735) | 13663..13896 | - | 234 | WP_130666525 | EexN family lipoprotein | - |
| ELH03_RS23740 (ELH03_23740) | 13893..14591 | - | 699 | WP_130766742 | P-type DNA transfer protein VirB5 | virB5 |
| ELH03_RS23745 (ELH03_23745) | 14591..16957 | - | 2367 | WP_130766743 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| ELH03_RS23750 (ELH03_23750) | 16950..17288 | - | 339 | WP_130766744 | type IV secretion system protein VirB3 | virB3 |
| ELH03_RS23755 (ELH03_23755) | 17294..17593 | - | 300 | WP_130766745 | TrbC/VirB2 family protein | virB2 |
| ELH03_RS23760 (ELH03_23760) | 17590..18252 | - | 663 | WP_130766746 | lytic transglycosylase domain-containing protein | virB1 |
| ELH03_RS38020 (ELH03_23765) | 18252..18791 | - | 540 | WP_130766747 | hypothetical protein | - |
| ELH03_RS23770 (ELH03_23770) | 18883..19236 | + | 354 | WP_128404288 | MarR family transcriptional regulator | - |
| ELH03_RS23775 (ELH03_23775) | 19412..21187 | - | 1776 | WP_245504888 | adenylate/guanylate cyclase domain-containing protein | - |
| ELH03_RS23780 (ELH03_23780) | 21171..22169 | - | 999 | WP_130766748 | ABC transporter substrate-binding protein | - |
Host bacterium
| ID | 19531 | GenBank | NZ_SILG01000002 |
| Plasmid name | pSM51_Rh01 | Incompatibility group | - |
| Plasmid size | 841308 bp | Coordinate of oriT [Strand] | 802876..802904 [-] |
| Host baterium | Rhizobium beringeri strain SM51 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | - |