Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 119056 |
| Name | oriT_pSO87_003 |
| Organism | Shigella sonnei strain ST152 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP140621 (27316..27407 [-], 92 nt) |
| oriT length | 92 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 92 nt
>oriT_pSO87_003
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTTTCAGGC
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTTTCAGGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file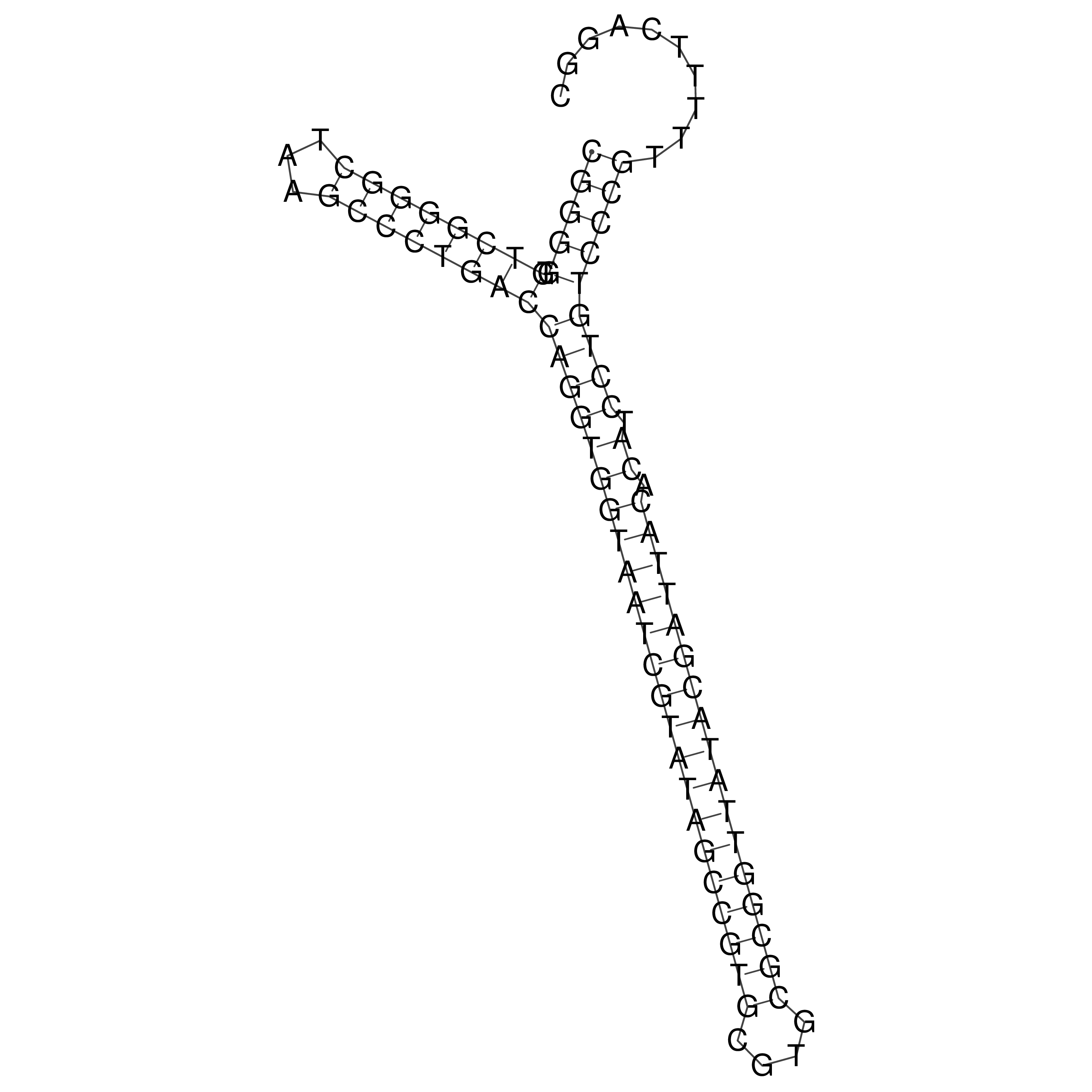
Relaxase
| ID | 12037 | GenBank | WP_000991405 |
| Name | Relaxase_O5Y90_RS25680_pSO87_003 |
UniProt ID | A0A896Z1V0 |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 104972.66 Da Isoelectric Point: 8.2349
>WP_000991405.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAAELAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIDQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDKAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAAELAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIDQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDKAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A896Z1V0 |
Auxiliary protein
| ID | 6873 | GenBank | WP_001291057 |
| Name | WP_001291057_pSO87_003 |
UniProt ID | A0A142CND4 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12596.69 Da Isoelectric Point: 10.7335
>WP_001291057.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRSGSEKRQKNVLIAVRFSPEEAEIVKEKAEKNGLTVSTLIRKTVLGKQINARIDEDFLKELMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITELANTLRRNLMGR
MSEKKTRSGSEKRQKNVLIAVRFSPEEAEIVKEKAEKNGLTVSTLIRKTVLGKQINARIDEDFLKELMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITELANTLRRNLMGR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CND4 |
T4CP
| ID | 14097 | GenBank | WP_013307869 |
| Name | trbC_O5Y90_RS25685_pSO87_003 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 86950.82 Da Isoelectric Point: 6.5117
>WP_013307869.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHVM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKVILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHVM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKVILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 32988..76102
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| O5Y90_RS25685 (O5Y90_25680) | 30707..33007 | - | 2301 | WP_013307869 | F-type conjugative transfer protein TrbC | - |
| O5Y90_RS25690 (O5Y90_25685) | 32988..34112 | - | 1125 | WP_000152382 | DsbC family protein | trbB |
| O5Y90_RS25695 (O5Y90_25690) | 34109..35452 | - | 1344 | WP_032253916 | hypothetical protein | trbA |
| O5Y90_RS25700 (O5Y90_25695) | 36249..36344 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| O5Y90_RS25705 (O5Y90_25700) | 36407..36583 | - | 177 | WP_001054907 | hypothetical protein | - |
| O5Y90_RS25710 (O5Y90_25705) | 36733..37200 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| O5Y90_RS25715 (O5Y90_25710) | 37359..37982 | + | 624 | WP_001155247 | helix-turn-helix domain-containing protein | - |
| O5Y90_RS25720 (O5Y90_25715) | 38180..38395 | - | 216 | WP_000266543 | hypothetical protein | - |
| O5Y90_RS25725 (O5Y90_25720) | 38399..38767 | - | 369 | WP_001345852 | hypothetical protein | - |
| O5Y90_RS25730 (O5Y90_25725) | 38779..38970 | - | 192 | WP_000751583 | hypothetical protein | - |
| O5Y90_RS25735 (O5Y90_25730) | 39058..39300 | - | 243 | WP_000650923 | hypothetical protein | - |
| O5Y90_RS25740 (O5Y90_25735) | 39630..39782 | + | 153 | WP_001302699 | Hok/Gef family protein | - |
| O5Y90_RS25745 (O5Y90_25740) | 39920..41602 | - | 1683 | WP_323739365 | TnsA endonuclease N-terminal domain-containing protein | - |
| O5Y90_RS25750 (O5Y90_25745) | 41522..42218 | + | 697 | WP_094110542 | IS1 family transposase | - |
| O5Y90_RS25755 (O5Y90_25750) | 42556..43209 | - | 654 | WP_001401768 | plasmid IncI1-type surface exclusion protein ExcA | - |
| O5Y90_RS25760 (O5Y90_25755) | 43297..45462 | - | 2166 | WP_000691808 | DotA/TraY family protein | traY |
| O5Y90_RS25765 (O5Y90_25760) | 45537..46106 | - | 570 | WP_000650752 | conjugal transfer protein TraX | - |
| O5Y90_RS25770 (O5Y90_25765) | 46103..47308 | - | 1206 | WP_001189548 | conjugal transfer protein TraW | traW |
| O5Y90_RS25775 (O5Y90_25770) | 47266..47886 | - | 621 | WP_000286854 | hypothetical protein | traV |
| O5Y90_RS25780 (O5Y90_25775) | 47886..50930 | - | 3045 | WP_001024766 | conjugal transfer protein | traU |
| O5Y90_RS25785 (O5Y90_25780) | 51225..51851 | - | 627 | WP_000785703 | hypothetical protein | traT |
| O5Y90_RS25790 (O5Y90_25785) | 51958..52242 | - | 285 | WP_001353683 | hypothetical protein | - |
| O5Y90_RS25795 (O5Y90_25790) | 52266..52664 | - | 399 | WP_000088812 | DUF6750 family protein | traR |
| O5Y90_RS25800 (O5Y90_25795) | 52711..53241 | - | 531 | WP_000987023 | conjugal transfer protein TraQ | traQ |
| O5Y90_RS25805 (O5Y90_25800) | 53238..53951 | - | 714 | WP_000787003 | conjugal transfer protein TraP | traP |
| O5Y90_RS25810 (O5Y90_25805) | 53948..55285 | - | 1338 | WP_013307873 | conjugal transfer protein TraO | traO |
| O5Y90_RS25815 (O5Y90_25810) | 55289..56263 | - | 975 | WP_000547383 | DotH/IcmK family type IV secretion protein | traN |
| O5Y90_RS25820 (O5Y90_25815) | 56274..56969 | - | 696 | WP_001681668 | DotI/IcmL family type IV secretion protein | traM |
| O5Y90_RS25825 (O5Y90_25820) | 56981..57331 | - | 351 | WP_001056367 | hypothetical protein | traL |
| O5Y90_RS25830 (O5Y90_25825) | 57348..61409 | - | 4062 | WP_013307874 | LPD7 domain-containing protein | - |
| O5Y90_RS25835 (O5Y90_25830) | 61763..62344 | - | 582 | WP_025750394 | hypothetical protein | - |
| O5Y90_RS25840 (O5Y90_25835) | 62576..62866 | - | 291 | WP_000817801 | IcmT/TraK family protein | traK |
| O5Y90_RS25845 (O5Y90_25840) | 62863..64011 | - | 1149 | WP_013307876 | plasmid transfer ATPase TraJ | virB11 |
| O5Y90_RS25850 (O5Y90_25845) | 63995..64831 | - | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| O5Y90_RS25855 (O5Y90_25850) | 64828..65286 | - | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| O5Y90_RS25860 (O5Y90_25855) | 65390..66592 | - | 1203 | WP_013307879 | conjugal transfer protein TraF | - |
| O5Y90_RS25865 (O5Y90_25860) | 66694..67515 | - | 822 | WP_001681675 | hypothetical protein | traE |
| O5Y90_RS25870 (O5Y90_25865) | 67728..68852 | - | 1125 | WP_013307880 | site-specific integrase | - |
| O5Y90_RS25875 (O5Y90_25870) | 68916..69305 | + | 390 | Protein_77 | phage tail protein | - |
| O5Y90_RS25880 (O5Y90_25875) | 69310..69549 | - | 240 | WP_000271302 | hypothetical protein | - |
| O5Y90_RS25885 (O5Y90_25880) | 70475..71710 | - | 1236 | WP_085008799 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| O5Y90_RS25890 (O5Y90_25885) | 71728..72354 | - | 627 | WP_013307883 | prepilin peptidase | - |
| O5Y90_RS25895 (O5Y90_25890) | 72370..72855 | - | 486 | WP_001080767 | lytic transglycosylase domain-containing protein | virB1 |
| O5Y90_RS25900 (O5Y90_25895) | 72900..73436 | - | 537 | WP_000111521 | type 4 pilus major pilin | - |
| O5Y90_RS25905 (O5Y90_25900) | 73498..74592 | - | 1095 | WP_000697154 | type II secretion system F family protein | - |
| O5Y90_RS25910 (O5Y90_25905) | 74594..76102 | - | 1509 | WP_000880806 | GspE/PulE family protein | virB11 |
| O5Y90_RS25915 (O5Y90_25910) | 76205..76663 | - | 459 | WP_000095887 | type IV pilus biogenesis protein PilP | - |
| O5Y90_RS25920 (O5Y90_25915) | 76653..77948 | - | 1296 | WP_000129885 | type 4b pilus protein PilO2 | - |
| O5Y90_RS25925 (O5Y90_25920) | 77969..79588 | - | 1620 | WP_001035562 | PilN family type IVB pilus formation outer membrane protein | - |
| O5Y90_RS25930 (O5Y90_25925) | 79620..80033 | - | 414 | WP_124983997 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 19488 | GenBank | NZ_CP140621 |
| Plasmid name | pSO87_003 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 86884 bp | Coordinate of oriT [Strand] | 27316..27407 [-] |
| Host baterium | Shigella sonnei strain ST152 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |