Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118962 |
| Name | oriT_pAF20192 |
| Organism | Pediococcus acidilactici strain AF2019 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP138504 (2723..2760 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..7, 19..25 (ACACAAC..GTTGTGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pAF20192
ACACAACCAACTTTAGTTGTTGTGTGTAAGTGCGCATT
ACACAACCAACTTTAGTTGTTGTGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file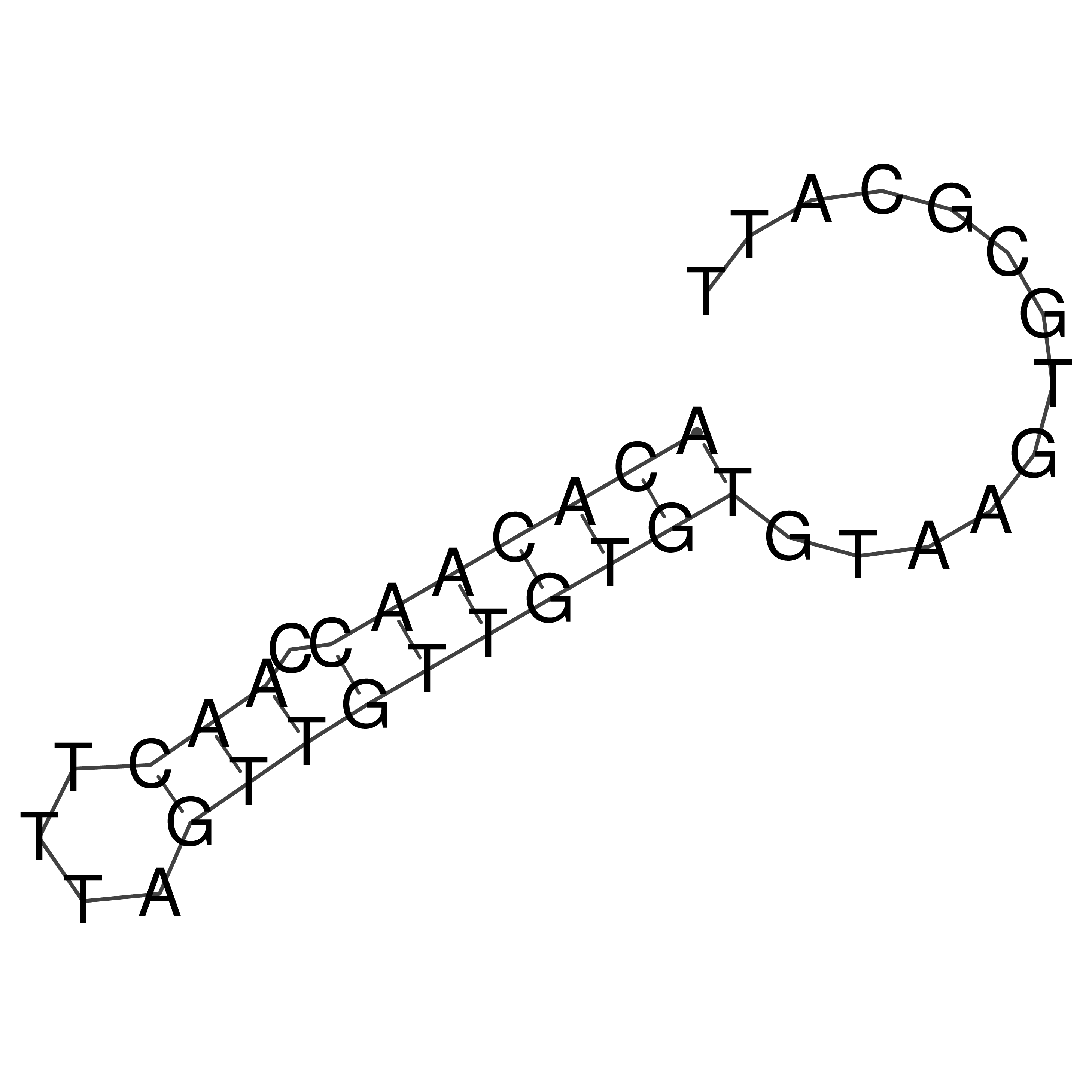
Relaxase
| ID | 12017 | GenBank | WP_317072632 |
| Name | mobQ_SGW13_RS10925_pAF20192 |
UniProt ID | _ |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80579.00 Da Isoelectric Point: 9.4248
>WP_317072632.1 MobQ family relaxase [Pediococcus acidilactici]
MAIFHMSFSNISAGKGRSAIAGASYRSGEKLFDQKEGRSYFYARSVMPESFILTPKNAPAWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNPDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKITEWRHNWAASVNQVLEQKNIP
DRISEKSFIEQGIDDTPTQHEGINSKRHERKEFNQQVKNYRKTKASYKNNQEKVINRGHLDSLSKHFSFN
EKRVVNELSHELKTYISLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETESEQTVFKGSELNERLMNIRDDLLTQQLLTFTKRPYVGFKLL
MQQEKEVKIELKYTLMIHNDNLESLEHVDQGLLEKYSPTEQQKITRAVKDLRTIMAVKQVIQTQYHEVLK
RAFPKGDLDGLPLTKQEQAYTAVMYYDPTLKPLKVETMEQWQANPPQVFSTQEHQLGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFFGECKVDPTIKNSQIEKIQKQLKEQQAKDDQYRKANMGHYQPLNYKPVSPDY
YLKTAFSNAIMTALYARDEDYQRQKQAQGLKETEWEMTKKQRQHQTRNRHEDGGMHL
MAIFHMSFSNISAGKGRSAIAGASYRSGEKLFDQKEGRSYFYARSVMPESFILTPKNAPAWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNPDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKITEWRHNWAASVNQVLEQKNIP
DRISEKSFIEQGIDDTPTQHEGINSKRHERKEFNQQVKNYRKTKASYKNNQEKVINRGHLDSLSKHFSFN
EKRVVNELSHELKTYISLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETESEQTVFKGSELNERLMNIRDDLLTQQLLTFTKRPYVGFKLL
MQQEKEVKIELKYTLMIHNDNLESLEHVDQGLLEKYSPTEQQKITRAVKDLRTIMAVKQVIQTQYHEVLK
RAFPKGDLDGLPLTKQEQAYTAVMYYDPTLKPLKVETMEQWQANPPQVFSTQEHQLGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFFGECKVDPTIKNSQIEKIQKQLKEQQAKDDQYRKANMGHYQPLNYKPVSPDY
YLKTAFSNAIMTALYARDEDYQRQKQAQGLKETEWEMTKKQRQHQTRNRHEDGGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14077 | GenBank | WP_317072617 |
| Name | t4cp2_SGW13_RS10985_pAF20192 |
UniProt ID | _ |
| Length | 503 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 503 a.a. Molecular weight: 56698.89 Da Isoelectric Point: 5.5070
>WP_317072617.1 VirD4-like conjugal transfer protein, CD1115 family [Pediococcus acidilactici]
MNGTILGIVGKRIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRSPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVADFTSFSDFDLKEIGNDKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKEKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETHFKNKVDKWNQTVKAQHQAKKDDQTTDDEEDKLQDNI
DQELESRHKKMLG
MNGTILGIVGKRIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRSPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVADFTSFSDFDLKEIGNDKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKEKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETHFKNKVDKWNQTVKAQHQAKKDDQTTDDEEDKLQDNI
DQELESRHKKMLG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6314..16129
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SGW13_RS10900 (SGW13_10900) | 1400..1681 | + | 282 | WP_003660204 | type II toxin-antitoxin system RelB/DinJ family antitoxin | - |
| SGW13_RS10905 (SGW13_10905) | 1728..1913 | + | 186 | WP_152725677 | hypothetical protein | - |
| SGW13_RS10910 (SGW13_10910) | 1934..2074 | + | 141 | Protein_3 | ATPase | - |
| SGW13_RS10915 (SGW13_10915) | 2064..2342 | - | 279 | WP_014216278 | hypothetical protein | - |
| SGW13_RS10920 (SGW13_10920) | 2365..2592 | - | 228 | WP_003555660 | hypothetical protein | - |
| SGW13_RS10925 (SGW13_10925) | 2845..4908 | + | 2064 | WP_317072632 | MobQ family relaxase | - |
| SGW13_RS10930 (SGW13_10930) | 4992..5303 | + | 312 | WP_063846748 | hypothetical protein | - |
| SGW13_RS10935 (SGW13_10935) | 5339..5953 | + | 615 | Protein_8 | hypothetical protein | - |
| SGW13_RS10940 (SGW13_10940) | 5955..6293 | + | 339 | WP_317072629 | CagC family type IV secretion system protein | - |
| SGW13_RS10945 (SGW13_10945) | 6314..6676 | + | 363 | WP_014216295 | hypothetical protein | trsC |
| SGW13_RS10950 (SGW13_10950) | 6645..7304 | + | 660 | WP_317072626 | TrsD/TraD family conjugative transfer protein | trsD |
| SGW13_RS10955 (SGW13_10955) | 7316..9331 | + | 2016 | WP_317072624 | helicase HerA domain-containing protein | virb4 |
| SGW13_RS10960 (SGW13_10960) | 9324..10742 | + | 1419 | WP_210697269 | type VII secretion protein EssB/YukC | trsF |
| SGW13_RS10965 (SGW13_10965) | 10743..11897 | + | 1155 | WP_154962721 | phage tail tip lysozyme | orf14 |
| SGW13_RS10970 (SGW13_10970) | 11911..12528 | + | 618 | WP_025085235 | hypothetical protein | - |
| SGW13_RS10975 (SGW13_10975) | 12515..12883 | + | 369 | WP_064658633 | thioredoxin family protein | - |
| SGW13_RS10980 (SGW13_10980) | 12884..13354 | + | 471 | WP_317072619 | conjugal transfer protein | trsJ |
| SGW13_RS10985 (SGW13_10985) | 13356..14867 | + | 1512 | WP_317072617 | VirD4-like conjugal transfer protein, CD1115 family | - |
| SGW13_RS10990 (SGW13_10990) | 14882..15271 | + | 390 | WP_057961950 | hypothetical protein | - |
| SGW13_RS10995 (SGW13_10995) | 15290..16129 | + | 840 | WP_317072612 | conjugal transfer protein TrbL family protein | trsL |
| SGW13_RS11000 (SGW13_11000) | 16144..16554 | + | 411 | WP_317072610 | hypothetical protein | - |
| SGW13_RS11005 (SGW13_11005) | 16551..17036 | + | 486 | Protein_22 | toprim domain-containing protein | - |
| SGW13_RS11010 (SGW13_11010) | 17029..17646 | - | 618 | Protein_23 | DNA topoisomerase | - |
| SGW13_RS11015 (SGW13_11015) | 17656..18534 | - | 879 | Protein_24 | IS982 family transposase | - |
| SGW13_RS11020 (SGW13_11020) | 18731..18817 | + | 87 | Protein_25 | KxYKxGKxW signal peptide domain-containing protein | - |
| SGW13_RS11025 (SGW13_11025) | 18860..19690 | + | 831 | WP_323052506 | hypothetical protein | - |
Host bacterium
| ID | 19394 | GenBank | NZ_CP138504 |
| Plasmid name | pAF20192 | Incompatibility group | - |
| Plasmid size | 28691 bp | Coordinate of oriT [Strand] | 2723..2760 [-] |
| Host baterium | Pediococcus acidilactici strain AF2019 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |