Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118947 |
| Name | oriT_pAtK224a |
| Organism | Agrobacterium tumefaciens strain K224 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP115926 (207887..207915 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pAtK224a
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file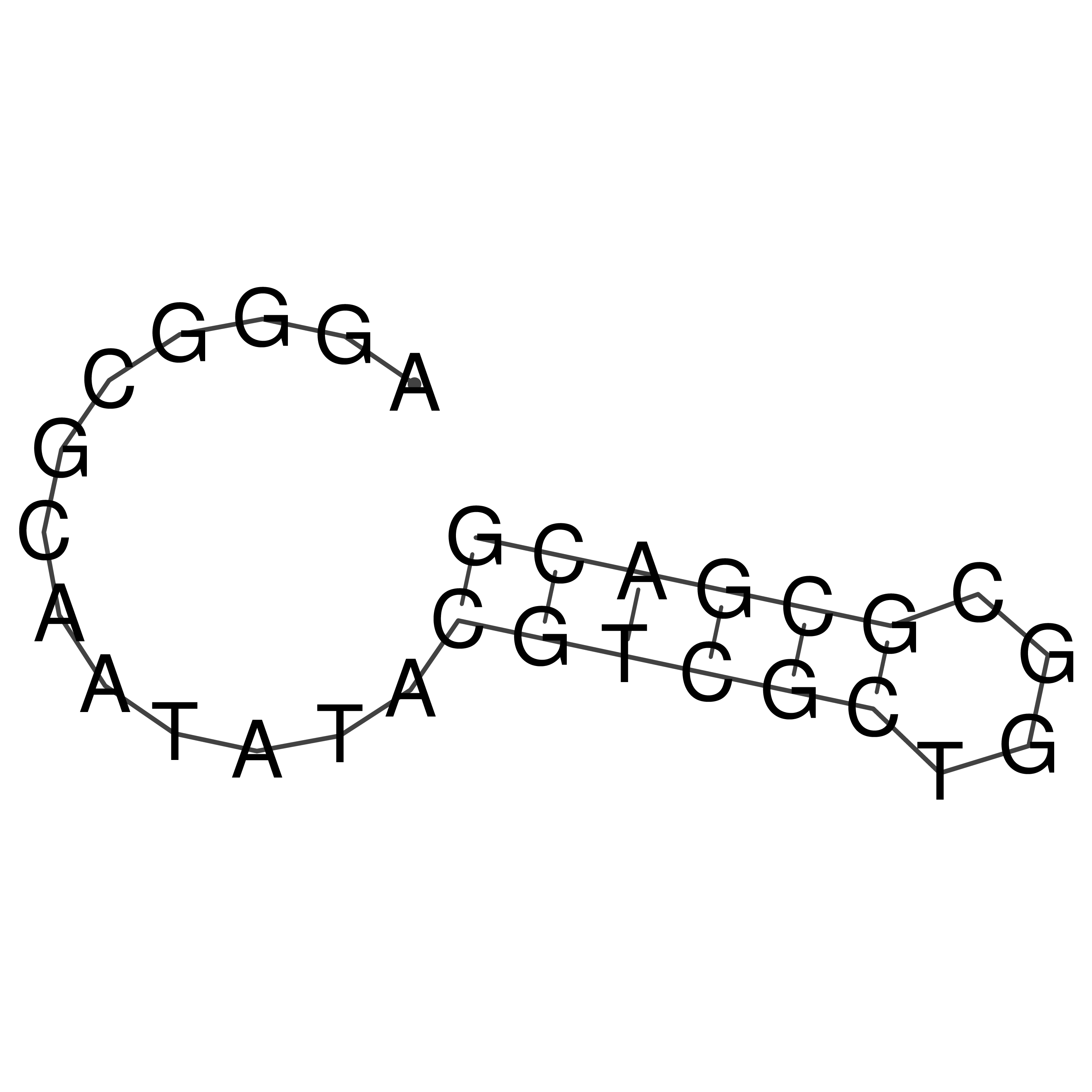
Relaxase
| ID | 12007 | GenBank | WP_173994836 |
| Name | traA_G6L41_RS23800_pAtK224a |
UniProt ID | _ |
| Length | 1542 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1542 a.a. Molecular weight: 171272.01 Da Isoelectric Point: 8.7441
>WP_173994836.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAAELKHEELALPDQVPAWLRTAIDG
KSVVAASEALWNAVDAFETRADAQLARELIIALPEELMRSENIALVREFVRDNLTSKGMIADWVYHDRQG
NPHIHLMTALRPLTQDGFGPKKVPVLGADGEPLRVVTPDRPKGKIVYRLWAGDKETMKSWKIAWADTANR
HLALAGHDIRLDGRSYVEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLADPYLLLKQ
LGNERSTFDERDIAKALHRYVDDPADFANIRAKLMASNDLVLLKPQQANLQTGKVAEPAVFTTSDILRTE
YDMAQSAEVLARRRGLGVAAAKIAAAVRTIETANPENLFTLDREQVEAVHHVAGDTGIAAVVGLAGAGKS
TLLAAARVAWESDNHRVFGAALAGKAAEGLEDSSGIRSRTLAYWELAWDNGRDLLDRRDIFVIDEAGMVS
SQQMARVLKRAEEAGAKVVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQREEWAREASRLFACGN
VKEGLDAYAQRGRIVEAESRAEAVERIVADWTSARRELIQQHADKEQPVRLRGDELLVLAHTNDDVGQLN
EALRNVMKDEGALTVSREFQTERGVREFAVGDRIIFLENARFLEPRAQRLGPQYVKNGMLGTVVSTGDTR
GGPLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRADL
YAAREDFEPKPEWEKRSRADHAAGITGELVETGNAKFRPQEEDADQSPYADVKTDDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERMTVTVSVIDEKTGEKRFEGRTVDRNVWTAMQIETAEVRRQRIEEESHRP
ELFKQLVERLGRSGAKTTSLDFESEAGYQAHARDFAKRRGIDTLSGVAAWMEDGAARQLAWLAQKREQVA
KHWERASVALGFAIESEKRVSYSEERAETGTAKIPTDRQYLIPPTTLFFRGVGEDARRAQLRSDHWKERD
AILRPVLQKIYRDPEGALARLNTLASDARIAPRELADDLEASPQRLGRLHGSDLLVDGRAARSERDAAVS
ALSELLPLARAHATEFRRQVEHFGMREEQRRAHMSLSIPALSKPAMARLAQIEAVSERGGKDAYKTAFAY
AAEHRLFVQEVRGVNDALNARFGWSAFTDKADTVAQRNMIERMPDDLDLDRRDKLNRLFTAIRRFAEEQH
LAERKDRSRIVTGASVVPGKETVPVLPMLAAVMEFETPIDHEARERARAVPHYSQNRAALVNAAARIWRD
PAGAVGKIEDLILKGFAGERIAAAISNDPAAYGALRGSGRIMDKLLAVGLERREAFRALPEAASRVRSLA
TSFRAAFDAEREAINEERRRMSIAIPGLSPSAEDALRELSVAMKKKNGRLDVAAGSLDPAISKEFATVSR
ALDERFGRNAILRGELDVINRVPPAQRRAFEVMREQLKVLQQVVRMQASQNIVAERHRRVLDRARGVILF
NQ
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAAELKHEELALPDQVPAWLRTAIDG
KSVVAASEALWNAVDAFETRADAQLARELIIALPEELMRSENIALVREFVRDNLTSKGMIADWVYHDRQG
NPHIHLMTALRPLTQDGFGPKKVPVLGADGEPLRVVTPDRPKGKIVYRLWAGDKETMKSWKIAWADTANR
HLALAGHDIRLDGRSYVEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLADPYLLLKQ
LGNERSTFDERDIAKALHRYVDDPADFANIRAKLMASNDLVLLKPQQANLQTGKVAEPAVFTTSDILRTE
YDMAQSAEVLARRRGLGVAAAKIAAAVRTIETANPENLFTLDREQVEAVHHVAGDTGIAAVVGLAGAGKS
TLLAAARVAWESDNHRVFGAALAGKAAEGLEDSSGIRSRTLAYWELAWDNGRDLLDRRDIFVIDEAGMVS
SQQMARVLKRAEEAGAKVVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQREEWAREASRLFACGN
VKEGLDAYAQRGRIVEAESRAEAVERIVADWTSARRELIQQHADKEQPVRLRGDELLVLAHTNDDVGQLN
EALRNVMKDEGALTVSREFQTERGVREFAVGDRIIFLENARFLEPRAQRLGPQYVKNGMLGTVVSTGDTR
GGPLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRADL
YAAREDFEPKPEWEKRSRADHAAGITGELVETGNAKFRPQEEDADQSPYADVKTDDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERMTVTVSVIDEKTGEKRFEGRTVDRNVWTAMQIETAEVRRQRIEEESHRP
ELFKQLVERLGRSGAKTTSLDFESEAGYQAHARDFAKRRGIDTLSGVAAWMEDGAARQLAWLAQKREQVA
KHWERASVALGFAIESEKRVSYSEERAETGTAKIPTDRQYLIPPTTLFFRGVGEDARRAQLRSDHWKERD
AILRPVLQKIYRDPEGALARLNTLASDARIAPRELADDLEASPQRLGRLHGSDLLVDGRAARSERDAAVS
ALSELLPLARAHATEFRRQVEHFGMREEQRRAHMSLSIPALSKPAMARLAQIEAVSERGGKDAYKTAFAY
AAEHRLFVQEVRGVNDALNARFGWSAFTDKADTVAQRNMIERMPDDLDLDRRDKLNRLFTAIRRFAEEQH
LAERKDRSRIVTGASVVPGKETVPVLPMLAAVMEFETPIDHEARERARAVPHYSQNRAALVNAAARIWRD
PAGAVGKIEDLILKGFAGERIAAAISNDPAAYGALRGSGRIMDKLLAVGLERREAFRALPEAASRVRSLA
TSFRAAFDAEREAINEERRRMSIAIPGLSPSAEDALRELSVAMKKKNGRLDVAAGSLDPAISKEFATVSR
ALDERFGRNAILRGELDVINRVPPAQRRAFEVMREQLKVLQQVVRMQASQNIVAERHRRVLDRARGVILF
NQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 14059 | GenBank | WP_173994838 |
| Name | traG_G6L41_RS23785_pAtK224a |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70732.67 Da Isoelectric Point: 8.9948
>WP_173994838.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MKGKAKQHPSLLLITIPIAVTGFSFYVAHWRWPELATGITGKTQYWFLRASPVPVLLFGPLAGLLAVWAL
PLHRRRPVALASFLCFLLMAGFYGMREFGRLQPLVESGVLTWDRALSFVDMIAVVGAAMGFMAAAVAARI
SSVVPEQVTRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPNDPVTWGRGGKAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYTGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESLRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSASAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFILDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWSAKNSASRKSENINFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMHAR
PNRFAKLGP
MKGKAKQHPSLLLITIPIAVTGFSFYVAHWRWPELATGITGKTQYWFLRASPVPVLLFGPLAGLLAVWAL
PLHRRRPVALASFLCFLLMAGFYGMREFGRLQPLVESGVLTWDRALSFVDMIAVVGAAMGFMAAAVAARI
SSVVPEQVTRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPNDPVTWGRGGKAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYTGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESLRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSASAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFILDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWSAKNSASRKSENINFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMHAR
PNRFAKLGP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 259751..269356
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G6L41_RS24030 (G6L41_024030) | 255209..256066 | + | 858 | WP_173994803 | SDR family oxidoreductase | - |
| G6L41_RS24035 (G6L41_024035) | 256171..256542 | + | 372 | WP_173994802 | DUF1330 domain-containing protein | - |
| G6L41_RS24040 (G6L41_024040) | 256741..257130 | - | 390 | WP_236762322 | hypothetical protein | - |
| G6L41_RS24045 (G6L41_024045) | 257310..257621 | - | 312 | WP_162633665 | hypothetical protein | - |
| G6L41_RS24050 (G6L41_024050) | 257988..258392 | - | 405 | WP_080802722 | host attachment protein | - |
| G6L41_RS24055 (G6L41_024055) | 258754..259116 | - | 363 | WP_080802725 | transcriptional regulator | - |
| G6L41_RS24060 (G6L41_024060) | 259210..259749 | + | 540 | WP_236762320 | hypothetical protein | - |
| G6L41_RS24065 (G6L41_024065) | 259751..260440 | + | 690 | WP_173994800 | type IV secretion system protein VirB1 | virB1 |
| G6L41_RS24070 (G6L41_024070) | 260437..260736 | + | 300 | WP_111790830 | TrbC/VirB2 family protein | virB2 |
| G6L41_RS24075 (G6L41_024075) | 260739..261080 | + | 342 | WP_013637408 | type IV secretion system protein VirB3 | virB3 |
| G6L41_RS24080 (G6L41_024080) | 261073..263454 | + | 2382 | WP_173994799 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| G6L41_RS24085 (G6L41_024085) | 263454..264152 | + | 699 | WP_173994798 | P-type DNA transfer protein VirB5 | virB5 |
| G6L41_RS24090 (G6L41_024090) | 264149..264382 | + | 234 | WP_003517253 | EexN family lipoprotein | - |
| G6L41_RS24095 (G6L41_024095) | 264387..265322 | + | 936 | WP_003517252 | type IV secretion system protein | virB6 |
| G6L41_RS24100 (G6L41_024100) | 265358..265624 | + | 267 | WP_003517250 | hypothetical protein | - |
| G6L41_RS24105 (G6L41_024105) | 265628..266299 | + | 672 | WP_173994797 | virB8 family protein | virB8 |
| G6L41_RS24110 (G6L41_024110) | 266299..267150 | + | 852 | WP_173994860 | P-type conjugative transfer protein VirB9 | virB9 |
| G6L41_RS24115 (G6L41_024115) | 267165..268337 | + | 1173 | WP_173994796 | type IV secretion system protein VirB10 | virB10 |
| G6L41_RS24120 (G6L41_024120) | 268346..269356 | + | 1011 | WP_173994795 | P-type DNA transfer ATPase VirB11 | virB11 |
| G6L41_RS24125 (G6L41_024125) | 269699..269974 | - | 276 | WP_003517240 | helix-turn-helix transcriptional regulator | - |
| G6L41_RS24130 (G6L41_024130) | 270505..271464 | + | 960 | WP_013637417 | AraC family transcriptional regulator | - |
| G6L41_RS24135 (G6L41_024135) | 271878..272291 | - | 414 | WP_173994794 | nucleoside diphosphate kinase regulator | - |
| G6L41_RS24140 (G6L41_024140) | 272811..273236 | + | 426 | WP_111790837 | GreA/GreB family elongation factor | - |
| G6L41_RS24145 (G6L41_024145) | 273419..274321 | + | 903 | WP_236762318 | DMT family transporter | - |
Host bacterium
| ID | 19379 | GenBank | NZ_CP115926 |
| Plasmid name | pAtK224a | Incompatibility group | - |
| Plasmid size | 485342 bp | Coordinate of oriT [Strand] | 207887..207915 [+] |
| Host baterium | Agrobacterium tumefaciens strain K224 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | PFL_4714 |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |