Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118693 |
| Name | oriT_pFF1003_1 |
| Organism | Klebsiella africana strain FF1003 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP059392 (60416..60465 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pFF1003_1
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file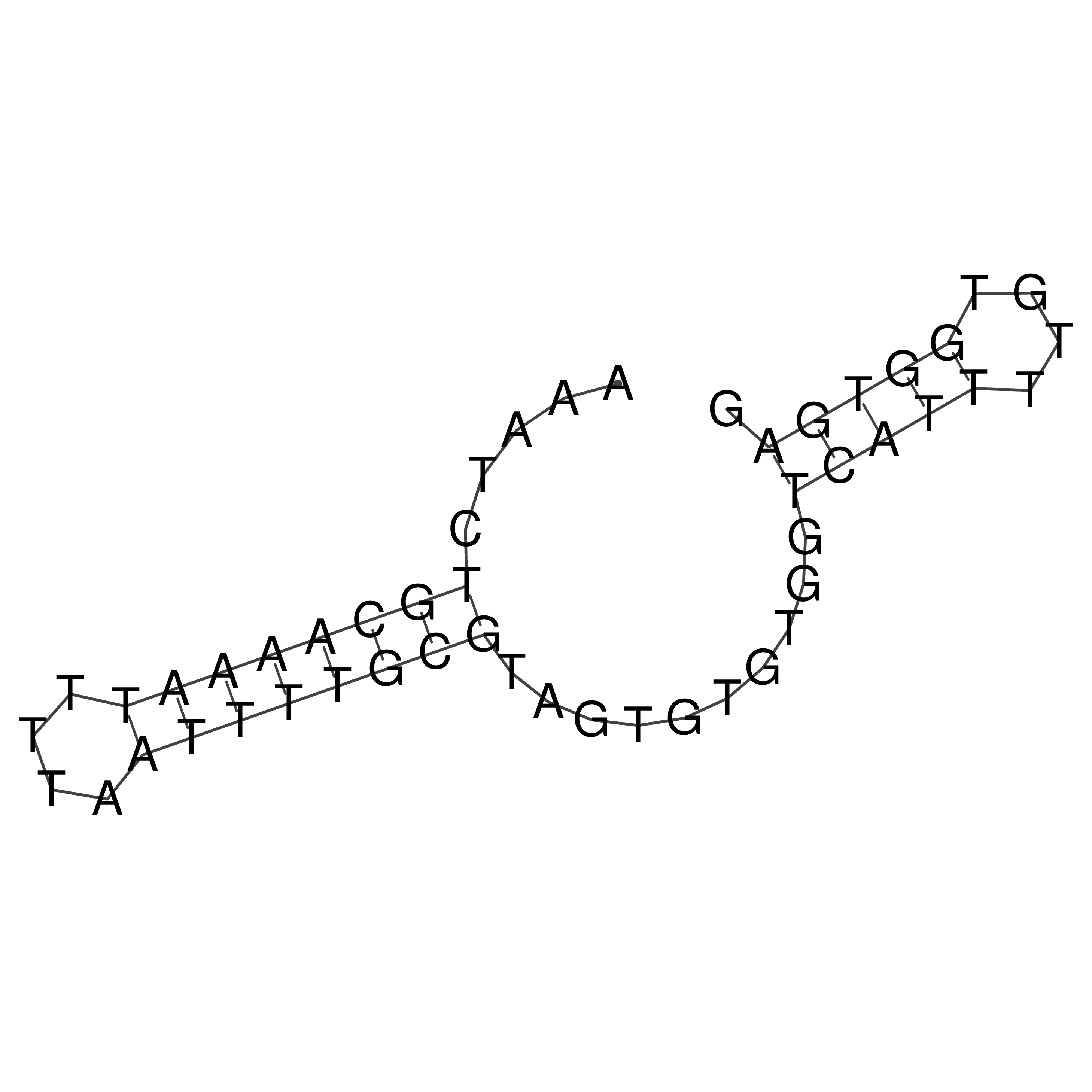
Relaxase
| ID | 11858 | GenBank | WP_203034335 |
| Name | traI_H1X61_RS25775_pFF1003_1 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190625.34 Da Isoelectric Point: 5.9980
>WP_203034335.1 conjugative transfer relaxase/helicase TraI [Klebsiella africana]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSLVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGAFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLHTPAEQAIHLSIPLLESQDLTFTRPQLLATALETGGDKVPMWSIEMTIK
KQIKSGQLLNVGVSPGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDIDRALATIEQVTPERVPRKEGAWVPGSSVVEFTPTQEEEIRK
ALSKGESLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNNDRRALNGLIHDARRENGETGREEITLP
VLVTSNIRDGELRKLSTWTAHKGAVALVDNVYHRIDNVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEEITVSQGDRMRFSKSDPERGYVANSVWEVQSVSGDSVTLSDGKTTRTLTPKADEAQQHIDLAYAITA
HGAQGASEPYAIALEGMVGARKLMASFESAYVGLSRMKQHVQVYTDRREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNIGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRDRLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSLVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGAFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLHTPAEQAIHLSIPLLESQDLTFTRPQLLATALETGGDKVPMWSIEMTIK
KQIKSGQLLNVGVSPGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDIDRALATIEQVTPERVPRKEGAWVPGSSVVEFTPTQEEEIRK
ALSKGESLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNNDRRALNGLIHDARRENGETGREEITLP
VLVTSNIRDGELRKLSTWTAHKGAVALVDNVYHRIDNVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEEITVSQGDRMRFSKSDPERGYVANSVWEVQSVSGDSVTLSDGKTTRTLTPKADEAQQHIDLAYAITA
HGAQGASEPYAIALEGMVGARKLMASFESAYVGLSRMKQHVQVYTDRREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNIGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRDRLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13889 | GenBank | WP_228290733 |
| Name | traD_H1X61_RS25770_pFF1003_1 |
UniProt ID | _ |
| Length | 760 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 760 a.a. Molecular weight: 85013.94 Da Isoelectric Point: 4.9596
>WP_228290733.1 type IV conjugative transfer system coupling protein TraD [Klebsiella africana]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDQSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTESHASEQPESVSPPA
PAEMTVTPAPVKAPPTTKRPAAEPPVLPVTPVPLLNQKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDQSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTESHASEQPESVSPPA
PAEMTVTPAPVKAPPTTKRPAAEPPVLPVTPVPLLNQKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 59858..88507
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H1X61_RS25555 (H1X61_25545) | 54875..55225 | + | 351 | WP_023320095 | hypothetical protein | - |
| H1X61_RS25560 (H1X61_25550) | 55861..56217 | + | 357 | WP_004152717 | hypothetical protein | - |
| H1X61_RS25565 (H1X61_25555) | 56278..56490 | + | 213 | WP_004182070 | hypothetical protein | - |
| H1X61_RS25570 (H1X61_25560) | 56501..56725 | + | 225 | WP_023320096 | hypothetical protein | - |
| H1X61_RS25575 (H1X61_25565) | 56806..57126 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| H1X61_RS25580 (H1X61_25570) | 57116..57394 | + | 279 | WP_203034326 | helix-turn-helix transcriptional regulator | - |
| H1X61_RS25585 (H1X61_25575) | 57395..57808 | + | 414 | WP_004182074 | type II toxin-antitoxin system HigA family antitoxin | - |
| H1X61_RS25590 (H1X61_25580) | 58642..59463 | + | 822 | WP_203034327 | DUF932 domain-containing protein | - |
| H1X61_RS25595 (H1X61_25585) | 59496..59825 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| H1X61_RS25600 (H1X61_25590) | 59858..60343 | - | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| H1X61_RS25605 (H1X61_25595) | 60734..61150 | + | 417 | WP_074181767 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| H1X61_RS25610 (H1X61_25600) | 61350..62069 | + | 720 | WP_203034328 | conjugal transfer protein TrbJ | - |
| H1X61_RS25615 (H1X61_25605) | 62202..62408 | + | 207 | WP_172412524 | TraY domain-containing protein | - |
| H1X61_RS25620 (H1X61_25610) | 62470..62838 | + | 369 | WP_064147976 | type IV conjugative transfer system pilin TraA | - |
| H1X61_RS25625 (H1X61_25615) | 62852..63157 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| H1X61_RS25630 (H1X61_25620) | 63177..63743 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| H1X61_RS25635 (H1X61_25625) | 63730..64470 | + | 741 | WP_203034329 | type-F conjugative transfer system secretin TraK | traK |
| H1X61_RS25640 (H1X61_25630) | 64470..65894 | + | 1425 | WP_016529134 | F-type conjugal transfer pilus assembly protein TraB | traB |
| H1X61_RS25645 (H1X61_25635) | 65887..66300 | + | 414 | Protein_78 | conjugal transfer pilus-stabilizing protein TraP | - |
| H1X61_RS25650 (H1X61_25640) | 66512..67081 | + | 570 | WP_015065628 | type IV conjugative transfer system lipoprotein TraV | traV |
| H1X61_RS26240 | 67237..67635 | + | 399 | WP_223177007 | hypothetical protein | - |
| H1X61_RS25660 (H1X61_25650) | 67664..67954 | + | 291 | WP_032426790 | hypothetical protein | - |
| H1X61_RS25665 (H1X61_25655) | 67978..68196 | + | 219 | WP_004171484 | hypothetical protein | - |
| H1X61_RS25670 (H1X61_25660) | 68197..68514 | + | 318 | WP_016530481 | hypothetical protein | - |
| H1X61_RS25675 (H1X61_25665) | 68581..68985 | + | 405 | WP_023320104 | hypothetical protein | - |
| H1X61_RS25680 (H1X61_25670) | 69010..69408 | + | 399 | WP_015065631 | hypothetical protein | - |
| H1X61_RS25685 (H1X61_25675) | 69480..72119 | + | 2640 | WP_203034330 | type IV secretion system protein TraC | virb4 |
| H1X61_RS25690 (H1X61_25680) | 72119..72508 | + | 390 | WP_203034331 | type-F conjugative transfer system protein TrbI | - |
| H1X61_RS25695 (H1X61_25685) | 72508..73134 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| H1X61_RS25700 (H1X61_25690) | 73176..73565 | + | 390 | WP_004194992 | hypothetical protein | - |
| H1X61_RS25705 (H1X61_25695) | 73562..74551 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| H1X61_RS25710 (H1X61_25700) | 74564..75202 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| H1X61_RS25715 (H1X61_25705) | 75261..77216 | + | 1956 | WP_203034332 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| H1X61_RS25720 (H1X61_25710) | 77248..77484 | + | 237 | WP_203034333 | conjugal transfer protein TrbE | - |
| H1X61_RS26355 | 77481..77666 | + | 186 | WP_032446653 | hypothetical protein | - |
| H1X61_RS25725 (H1X61_25715) | 77712..78038 | + | 327 | WP_016529362 | hypothetical protein | - |
| H1X61_RS25730 (H1X61_25720) | 78059..78811 | + | 753 | WP_203034334 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| H1X61_RS25735 (H1X61_25725) | 78822..79061 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| H1X61_RS25740 (H1X61_25730) | 79033..79590 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| H1X61_RS25745 (H1X61_25735) | 79635..80078 | + | 444 | WP_015065638 | F-type conjugal transfer protein TrbF | - |
| H1X61_RS25750 (H1X61_25740) | 80056..81435 | + | 1380 | WP_011977731 | conjugal transfer pilus assembly protein TraH | traH |
| H1X61_RS25755 (H1X61_25745) | 81435..84257 | + | 2823 | WP_023320108 | conjugal transfer mating-pair stabilization protein TraG | traG |
| H1X61_RS25760 (H1X61_25750) | 84281..84883 | + | 603 | WP_023280878 | hypothetical protein | - |
| H1X61_RS25765 (H1X61_25755) | 85134..85865 | + | 732 | WP_032415736 | conjugal transfer complement resistance protein TraT | - |
| H1X61_RS25770 (H1X61_25760) | 86225..88507 | + | 2283 | WP_228290733 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 19125 | GenBank | NZ_CP059392 |
| Plasmid name | pFF1003_1 | Incompatibility group | IncFIA |
| Plasmid size | 97925 bp | Coordinate of oriT [Strand] | 60416..60465 [-] |
| Host baterium | Klebsiella africana strain FF1003 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |