Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118501 |
| Name | oriT_pArA4 |
| Organism | Rhizobium rhizogenes strain A4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP073112 (160372..160405 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pArA4
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file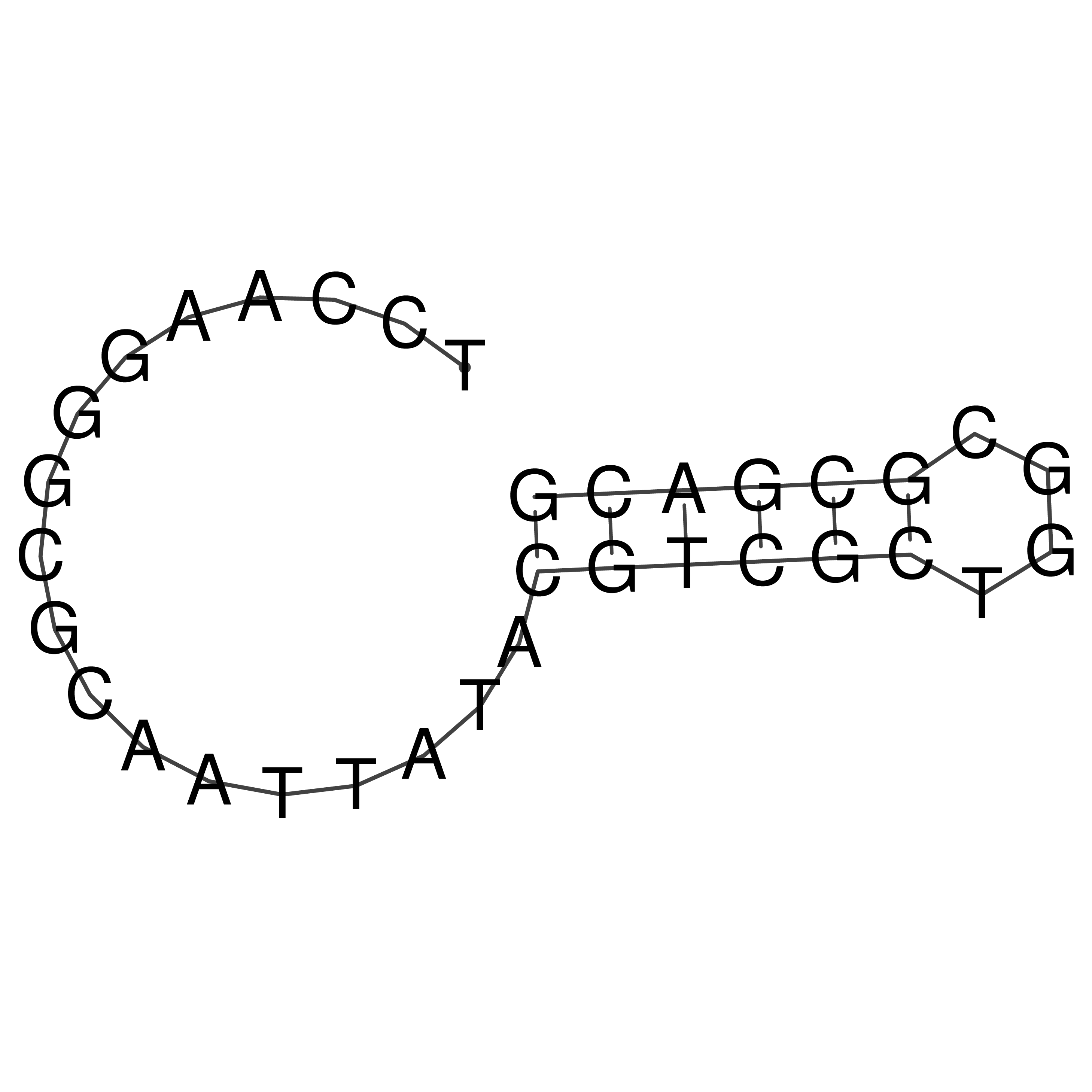
Relaxase
| ID | 11746 | GenBank | WP_034521179 |
| Name | traA_EML492_RS32565_pArA4 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123443.81 Da Isoelectric Point: 9.7034
>WP_034521179.1 Ti-type conjugative transfer relaxase TraA [Rhizobium rhizogenes]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELTAEQNIALMRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYDLWAGSTEEFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLELITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPVALRLERERIDFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHVRLSDEQRTAIEHVADGERIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNQLDDKTIIVLDEAGMVSSRQM
ALLVETVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNEMARAKLVERGIVANGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVLEAALGRIVAEIGEGEHRRKVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGDLVPILSRRNAKETTLDYE
KASFYGQALRFADTRGLHLMDVARTVARDRLEWTIRQSSRLVDLGVRLAAIATKLGFVGSFKNSPIQNPI
KEAKPMVSGITTFPKSLEQAVEDKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGVLASRADRQDREKAIANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQRAEVRNAWDAMRAVQQLAAHERTTKALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELTAEQNIALMRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYDLWAGSTEEFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLELITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPVALRLERERIDFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHVRLSDEQRTAIEHVADGERIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNQLDDKTIIVLDEAGMVSSRQM
ALLVETVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNEMARAKLVERGIVANGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVLEAALGRIVAEIGEGEHRRKVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGDLVPILSRRNAKETTLDYE
KASFYGQALRFADTRGLHLMDVARTVARDRLEWTIRQSSRLVDLGVRLAAIATKLGFVGSFKNSPIQNPI
KEAKPMVSGITTFPKSLEQAVEDKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGVLASRADRQDREKAIANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQRAEVRNAWDAMRAVQQLAAHERTTKALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13758 | GenBank | WP_212512283 |
| Name | traG_EML492_RS32550_pArA4 |
UniProt ID | _ |
| Length | 652 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 652 a.a. Molecular weight: 70234.40 Da Isoelectric Point: 9.4783
>WP_212512283.1 Ti-type conjugative transfer system protein TraG [Rhizobium rhizogenes]
MTVSRIALVAGPAALMILVVIGMTGIEQWLSDFGKTGAARLTWGRVGIALPYVVSSAIGILFLFASAGSI
NIKQAGWGALTGCTATILIAAIRETTRLGAFMKVLPADKTILAYLDPATSIGASAALLCACFALRVALIG
NAAFASAKPKRIHGKRALHGEADWMKLTEAATLFSDAGGIVIGERYRVDKDNVAAQAFRADNAETWGAGG
KSPLLCFNGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGSTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PKKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDE
TEQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGRKFSTSDIAGGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQLEGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VEIDQVSRSSQARGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAMWFRREAMKACVG
KNRFHGTRDPDATAQKAATRRL
MTVSRIALVAGPAALMILVVIGMTGIEQWLSDFGKTGAARLTWGRVGIALPYVVSSAIGILFLFASAGSI
NIKQAGWGALTGCTATILIAAIRETTRLGAFMKVLPADKTILAYLDPATSIGASAALLCACFALRVALIG
NAAFASAKPKRIHGKRALHGEADWMKLTEAATLFSDAGGIVIGERYRVDKDNVAAQAFRADNAETWGAGG
KSPLLCFNGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGSTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PKKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDE
TEQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGRKFSTSDIAGGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQLEGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VEIDQVSRSSQARGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAMWFRREAMKACVG
KNRFHGTRDPDATAQKAATRRL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 165543..180732
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EML492_RS32570 (EML492_32570) | 163837..164373 | + | 537 | WP_051696880 | conjugative transfer signal peptidase TraF | - |
| EML492_RS32575 (EML492_32575) | 164363..165526 | + | 1164 | WP_034521175 | conjugal transfer protein TraB | - |
| EML492_RS32580 (EML492_32580) | 165543..166133 | + | 591 | WP_034521172 | TraH family protein | virB1 |
| EML492_RS32585 (EML492_32585) | 166304..168157 | - | 1854 | WP_034521169 | zeta toxin family protein | - |
| EML492_RS32590 (EML492_32590) | 168391..169182 | - | 792 | WP_229202718 | hypothetical protein | - |
| EML492_RS32595 (EML492_32595) | 169447..169689 | - | 243 | WP_034521166 | helix-turn-helix domain-containing protein | - |
| EML492_RS32600 (EML492_32600) | 169885..170202 | + | 318 | WP_034521163 | transcriptional repressor TraM | - |
| EML492_RS32605 (EML492_32605) | 170195..170908 | - | 714 | WP_034521160 | autoinducer binding domain-containing protein | - |
| EML492_RS32610 (EML492_32610) | 171220..172518 | - | 1299 | WP_034521158 | IncP-type conjugal transfer protein TrbI | virB10 |
| EML492_RS32615 (EML492_32615) | 172530..172970 | - | 441 | WP_034521156 | conjugal transfer protein TrbH | - |
| EML492_RS32620 (EML492_32620) | 172974..173786 | - | 813 | WP_034521154 | P-type conjugative transfer protein TrbG | virB9 |
| EML492_RS32625 (EML492_32625) | 173804..174466 | - | 663 | WP_027668115 | conjugal transfer protein TrbF | virB8 |
| EML492_RS32630 (EML492_32630) | 174487..175674 | - | 1188 | WP_034521151 | P-type conjugative transfer protein TrbL | virB6 |
| EML492_RS32635 (EML492_32635) | 175679..175855 | - | 177 | WP_034521149 | entry exclusion protein TrbK | - |
| EML492_RS32640 (EML492_32640) | 175849..176652 | - | 804 | WP_034521147 | P-type conjugative transfer protein TrbJ | virB5 |
| EML492_RS32645 (EML492_32645) | 176624..179079 | - | 2456 | Protein_152 | conjugal transfer protein TrbE | - |
| EML492_RS32650 (EML492_32650) | 179090..179389 | - | 300 | WP_034521145 | conjugal transfer protein TrbD | virB3 |
| EML492_RS32655 (EML492_32655) | 179382..179765 | - | 384 | WP_034521141 | TrbC/VirB2 family protein | virB2 |
| EML492_RS32660 (EML492_32660) | 179755..180732 | - | 978 | WP_212512284 | P-type conjugative transfer ATPase TrbB | virB11 |
| EML492_RS32665 (EML492_32665) | 180744..181367 | - | 624 | WP_034523044 | acyl-homoserine-lactone synthase TraI | - |
Host bacterium
| ID | 18934 | GenBank | NZ_CP073112 |
| Plasmid name | pArA4 | Incompatibility group | - |
| Plasmid size | 181745 bp | Coordinate of oriT [Strand] | 160372..160405 [+] |
| Host baterium | Rhizobium rhizogenes strain A4 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |