Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118500 |
| Name | oriT_MSB1_1H|3 |
| Organism | Citrobacter freundii strain MSB1_1H isolate MSB1_1H |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR890183 (46484..46634 [+], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 12..17, 24..29 (AACCCG..CGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_MSB1_1H|3
TGGTCGATGTGAACCCGTTTGACCGGGTTATGAATGAGTTGAAAAGTCGTGGCCGCAAGAACGCTCATATCCTGAGCATCCTTCAATTCGACTGGCCCGCATCGGAGGTCATCATCGAGAAGCTGAGCTGTTACATCACAGATGGGATTAA
TGGTCGATGTGAACCCGTTTGACCGGGTTATGAATGAGTTGAAAAGTCGTGGCCGCAAGAACGCTCATATCCTGAGCATCCTTCAATTCGACTGGCCCGCATCGGAGGTCATCATCGAGAAGCTGAGCTGTTACATCACAGATGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file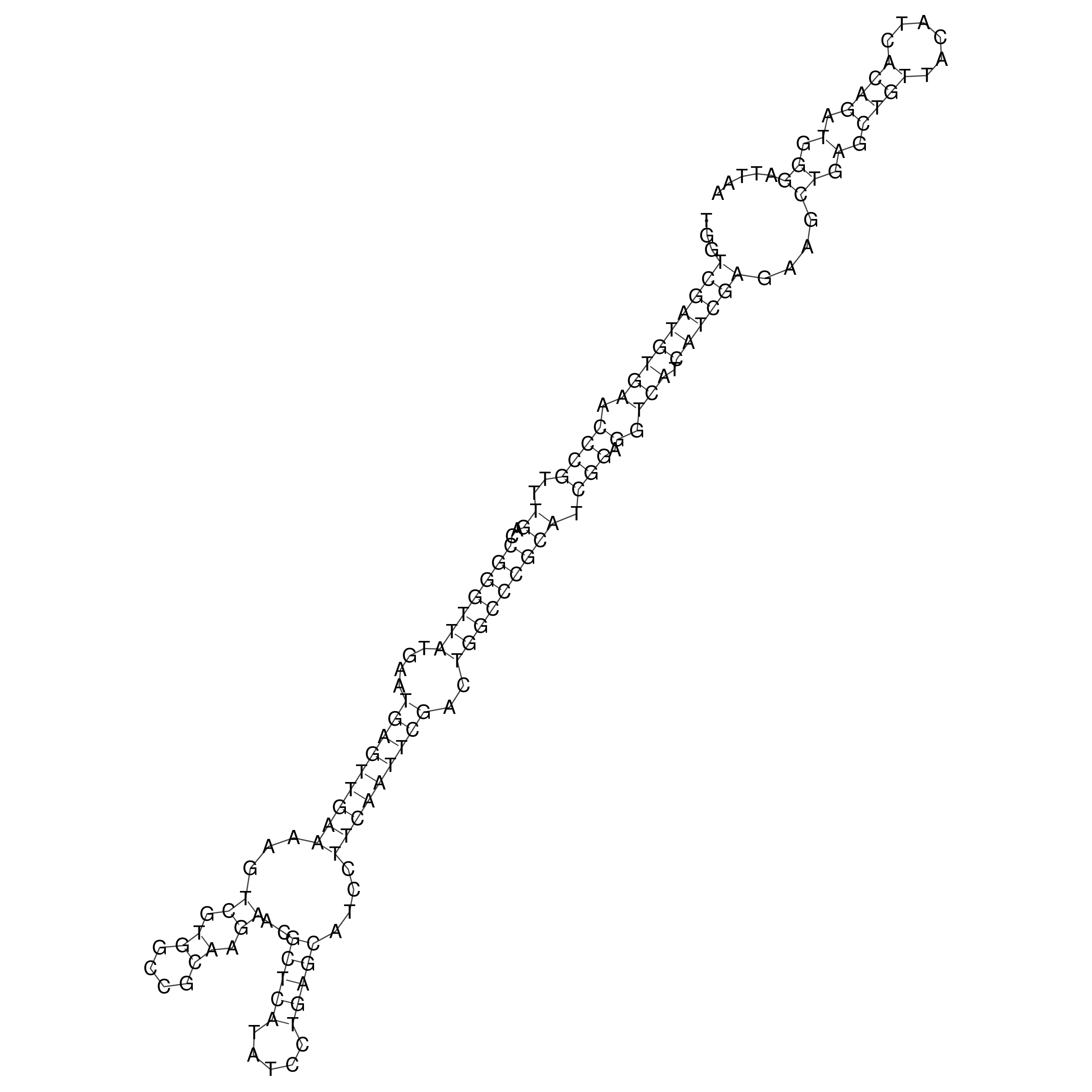
Relaxase
| ID | 11745 | GenBank | WP_000909947 |
| Name | mobH_JMV71_RS24630_MSB1_1H|3 |
UniProt ID | _ |
| Length | 987 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 987 a.a. Molecular weight: 109404.96 Da Isoelectric Point: 4.8922
>WP_000909947.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALHKLFGGRSGAIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAYRHGIDRYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVNEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGPVKILMLRLESNDLVFTTEPPAAVTGEVVGDVEDAEIE
FVDPEEADEDQAEGDAGLNNDMLAAELEAEKALAGLGFGDAMAMLKSTSDIAEEKPEQEDTESTELPKPD
ASKKGKQQSKPGKARAEKQPQKPEVKEDLSPQDIAKNAPPLANDDPLQALKDVGGGLGDIDFPFDVFNAS
AETACTDATNLEIPDITEPEKQEEQPKPDFVPQEQNSLQNGDFPIFSDSDEPPSWAIEPLPMLTDTPEQT
IPAPDMQHAGKPKELEKDAKTLLSEMLAGYGEASTLLEQAIMPVLEGKTTLGEVLCLMKGQAVILYPEGA
RSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLVDQLSDAVVAAIKDAEASMGGYQEAFELV
SPPGSGVSKNKSAPKQQSRKKTQQQKPEVDAGAGSEQKTKDKSLQQQPKEKQGNVASLVEEQKRKPVQDQ
EKQNVARLPKREAQPVAPKPKVEHERELGHVEVRERDEPEVREFEPPKAKTNPKDINEEDFLPSGVTPEK
ALLMLKDMIQKRSGRWLVTPVLEEDGCLVTSVKAFDMIAGENVGISKHILYGLLSRAQRRPLLKKRQGKL
YLEVDKA
MLKALHKLFGGRSGAIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAYRHGIDRYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVNEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGPVKILMLRLESNDLVFTTEPPAAVTGEVVGDVEDAEIE
FVDPEEADEDQAEGDAGLNNDMLAAELEAEKALAGLGFGDAMAMLKSTSDIAEEKPEQEDTESTELPKPD
ASKKGKQQSKPGKARAEKQPQKPEVKEDLSPQDIAKNAPPLANDDPLQALKDVGGGLGDIDFPFDVFNAS
AETACTDATNLEIPDITEPEKQEEQPKPDFVPQEQNSLQNGDFPIFSDSDEPPSWAIEPLPMLTDTPEQT
IPAPDMQHAGKPKELEKDAKTLLSEMLAGYGEASTLLEQAIMPVLEGKTTLGEVLCLMKGQAVILYPEGA
RSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLVDQLSDAVVAAIKDAEASMGGYQEAFELV
SPPGSGVSKNKSAPKQQSRKKTQQQKPEVDAGAGSEQKTKDKSLQQQPKEKQGNVASLVEEQKRKPVQDQ
EKQNVARLPKREAQPVAPKPKVEHERELGHVEVRERDEPEVREFEPPKAKTNPKDINEEDFLPSGVTPEK
ALLMLKDMIQKRSGRWLVTPVLEEDGCLVTSVKAFDMIAGENVGISKHILYGLLSRAQRRPLLKKRQGKL
YLEVDKA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13756 | GenBank | WP_000178858 |
| Name | traD_JMV71_RS24635_MSB1_1H|3 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69248.76 Da Isoelectric Point: 7.1093
>WP_000178858.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWMAASCAALAVEQVSTMPSEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVLTHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVKPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNTDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWMAASCAALAVEQVSTMPSEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVLTHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVKPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNTDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13757 | GenBank | WP_012766329 |
| Name | traC_JMV71_RS24715_MSB1_1H|3 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92529.82 Da Isoelectric Point: 5.8328
>WP_012766329.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIITIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDQSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTSLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRILAKTSKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQYLAQIRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDREAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYDDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSQKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSDGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRKQAA
MIITIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDQSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTSLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRILAKTSKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQYLAQIRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDREAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYDDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSQKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSDGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRKQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 44568..69672
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JMV71_RS24620 | 40608..40841 | - | 234 | WP_001191893 | hypothetical protein | - |
| JMV71_RS24625 | 40823..41440 | - | 618 | WP_012766323 | hypothetical protein | - |
| JMV71_RS24630 | 41608..44571 | + | 2964 | WP_000909947 | MobH family relaxase | - |
| JMV71_RS24635 | 44568..46433 | + | 1866 | WP_000178858 | conjugative transfer system coupling protein TraD | virb4 |
| JMV71_RS24640 | 46483..47028 | + | 546 | WP_023356350 | hypothetical protein | - |
| JMV71_RS24645 | 46985..47614 | + | 630 | WP_057521214 | DUF4400 domain-containing protein | tfc7 |
| JMV71_RS24650 | 47625..48071 | + | 447 | WP_057521213 | hypothetical protein | - |
| JMV71_RS24655 | 48081..48458 | + | 378 | WP_000869260 | hypothetical protein | - |
| JMV71_RS24660 | 48458..49120 | + | 663 | WP_057521212 | hypothetical protein | - |
| JMV71_RS24665 | 49299..49442 | + | 144 | WP_023356351 | hypothetical protein | - |
| JMV71_RS24670 | 49455..49802 | + | 348 | WP_001102160 | hypothetical protein | - |
| JMV71_RS24675 | 49945..50226 | + | 282 | WP_000805626 | type IV conjugative transfer system protein TraL | traL |
| JMV71_RS24680 | 50223..50849 | + | 627 | WP_057521211 | TraE/TraK family type IV conjugative transfer system protein | traE |
| JMV71_RS24685 | 50833..51750 | + | 918 | WP_012766326 | type-F conjugative transfer system secretin TraK | traK |
| JMV71_RS24690 | 51747..53063 | + | 1317 | WP_057521210 | TraB/VirB10 family protein | traB |
| JMV71_RS24695 | 53060..53638 | + | 579 | WP_039272410 | type IV conjugative transfer system lipoprotein TraV | traV |
| JMV71_RS24700 | 53642..54034 | + | 393 | WP_012766327 | TraA family conjugative transfer protein | - |
| JMV71_RS24705 | 54235..59721 | + | 5487 | WP_057521209 | Ig-like domain-containing protein | - |
| JMV71_RS24710 | 59870..60577 | + | 708 | WP_057521208 | DsbC family protein | - |
| JMV71_RS24715 | 60574..63021 | + | 2448 | WP_012766329 | type IV secretion system protein TraC | virb4 |
| JMV71_RS24720 | 63036..63353 | + | 318 | WP_000364398 | hypothetical protein | - |
| JMV71_RS24725 | 63350..63880 | + | 531 | WP_000672456 | S26 family signal peptidase | - |
| JMV71_RS24730 | 63843..65111 | + | 1269 | WP_057521207 | TrbC family F-type conjugative pilus assembly protein | traW |
| JMV71_RS24735 | 65108..65785 | + | 678 | WP_012766331 | EAL domain-containing protein | - |
| JMV71_RS24740 | 65779..66783 | + | 1005 | WP_000911304 | TraU family protein | traU |
| JMV71_RS24745 | 66934..69672 | + | 2739 | WP_057521206 | conjugal transfer mating pair stabilization protein TraN | traN |
| JMV71_RS24750 | 69711..70571 | - | 861 | WP_000679674 | hypothetical protein | - |
| JMV71_RS24755 | 70683..71333 | - | 651 | WP_000796641 | hypothetical protein | - |
| JMV71_RS24760 | 71624..71944 | + | 321 | WP_000547567 | hypothetical protein | - |
| JMV71_RS24765 | 72189..72440 | + | 252 | WP_226453752 | hypothetical protein | - |
| JMV71_RS24770 | 72661..73629 | + | 969 | WP_001553257 | AAA family ATPase | - |
| JMV71_RS24775 | 73641..74546 | + | 906 | WP_000739141 | hypothetical protein | - |
Host bacterium
| ID | 18933 | GenBank | NZ_LR890183 |
| Plasmid name | MSB1_1H|3 | Incompatibility group | IncA/C |
| Plasmid size | 126853 bp | Coordinate of oriT [Strand] | 46484..46634 [+] |
| Host baterium | Citrobacter freundii strain MSB1_1H isolate MSB1_1H |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |