Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118499 |
| Name | oriT_MSB1_1H|2 |
| Organism | Citrobacter freundii strain MSB1_1H isolate MSB1_1H |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR890182 (80862..80941 [-], 80 nt) |
| oriT length | 80 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 80 nt
>oriT_MSB1_1H|2
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCG
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file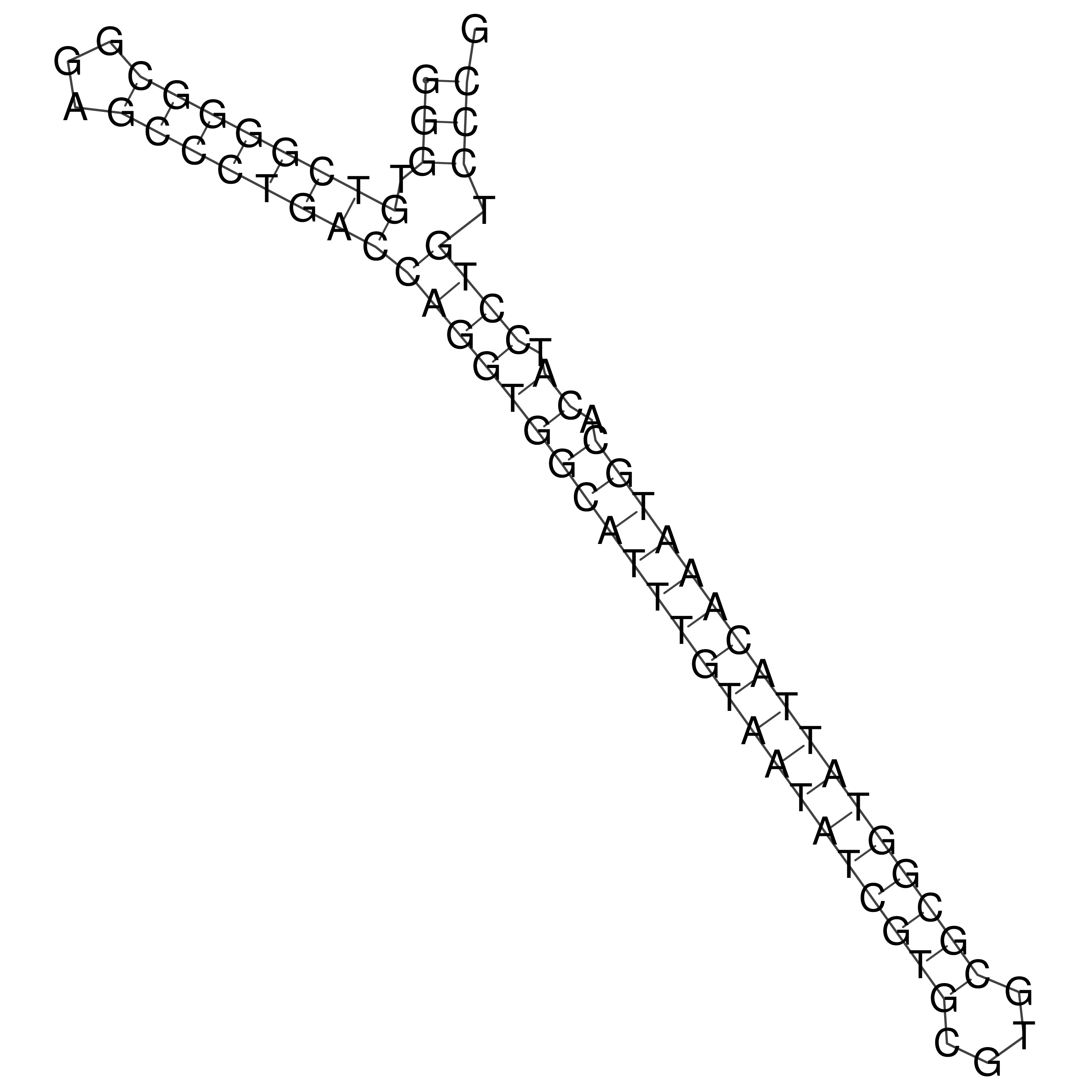
Relaxase
| ID | 11744 | GenBank | WP_048227654 |
| Name | Relaxase_JMV71_RS24075_MSB1_1H|2 |
UniProt ID | A0A8H9UN45 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102477.49 Da Isoelectric Point: 8.3859
>WP_048227654.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MIAVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVKASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGSEWVNFYGVTCFHNCTSIESAAEEMEFSARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVSAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSIERDRQSAWKRGRIQSLREYIADTAIAGLREAPVTDWSSLHQRFAKEGLFLSAD
NGELKVKDGWDSERPGVTLSAFGHAFDTEKLIRKLGEFTPPAQDIFSQVQAVGRYEPEKITVPSRPERMT
EKDTLTDYATARLRAPLIALDQDPSQRTIQSVHTLLAKSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAVLDSYKGGWKPVPKDIFQQVPPAEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQEIETLISSKHFSWERCHEQFAKQGLLLVREQQGLVVMDAYNREQTPVKASHVHPDLTLAR
AEPHAGSFVPVPADIFERVKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLRGRERYQAIHDECRMRKARIRIEHRDPLVRKLYYHIVELQRMQALITLKEAMKAERAELIGQGT
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYKDRRGNAVKSTTDSRCVVICEPGGTPMYSGVPGLQASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNTGDYDSLKRDMVKVAPVLFSRSPTVAMAPEGNDEQFNTAFAKM
VAWHNVNHRDEAEYHIARPDVDQLRVEHEQQFTNVLNQSSYAPDAQASHQNTWEPPTPR
MIAVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVKASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGSEWVNFYGVTCFHNCTSIESAAEEMEFSARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVSAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSIERDRQSAWKRGRIQSLREYIADTAIAGLREAPVTDWSSLHQRFAKEGLFLSAD
NGELKVKDGWDSERPGVTLSAFGHAFDTEKLIRKLGEFTPPAQDIFSQVQAVGRYEPEKITVPSRPERMT
EKDTLTDYATARLRAPLIALDQDPSQRTIQSVHTLLAKSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAVLDSYKGGWKPVPKDIFQQVPPAEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQEIETLISSKHFSWERCHEQFAKQGLLLVREQQGLVVMDAYNREQTPVKASHVHPDLTLAR
AEPHAGSFVPVPADIFERVKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLRGRERYQAIHDECRMRKARIRIEHRDPLVRKLYYHIVELQRMQALITLKEAMKAERAELIGQGT
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYKDRRGNAVKSTTDSRCVVICEPGGTPMYSGVPGLQASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNTGDYDSLKRDMVKVAPVLFSRSPTVAMAPEGNDEQFNTAFAKM
VAWHNVNHRDEAEYHIARPDVDQLRVEHEQQFTNVLNQSSYAPDAQASHQNTWEPPTPR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13755 | GenBank | WP_053389033 |
| Name | trbC_JMV71_RS24095_MSB1_1H|2 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87413.54 Da Isoelectric Point: 6.8253
>WP_053389033.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MNNTPVNQALMKRNAWNSPVLELMRDHLLYAGIMAGSVVAGFIWPLAIPLFLLLTIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSFFRAFPSLFQYETTQESKGKGIFYIGYKRMNDIGRELWLTLDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAGLYCRAVDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGKFPYMNYLDEVGAYYTERIAVQATQVRSLEFALVMMSQDQERIENQTSAANVATLM
QNAGTKVAGKIVSDDKTARTISNAAGQEARANMVSLQRQDGLIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAAENVSKIIG
VLTEKPSRRRKAARTEPWKIIDTFQQRLANRQEAHNLMTEYDMDHGGREENLWAEALEIIRTTTMAERTI
RYITLNKPESESTEVNAEEIQLAPDAILRNLTLPQPAYVPPARHRNEPFFSPKQTSSPQPDMEWPE
MNNTPVNQALMKRNAWNSPVLELMRDHLLYAGIMAGSVVAGFIWPLAIPLFLLLTIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSFFRAFPSLFQYETTQESKGKGIFYIGYKRMNDIGRELWLTLDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAGLYCRAVDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGKFPYMNYLDEVGAYYTERIAVQATQVRSLEFALVMMSQDQERIENQTSAANVATLM
QNAGTKVAGKIVSDDKTARTISNAAGQEARANMVSLQRQDGLIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAAENVSKIIG
VLTEKPSRRRKAARTEPWKIIDTFQQRLANRQEAHNLMTEYDMDHGGREENLWAEALEIIRTTTMAERTI
RYITLNKPESESTEVNAEEIQLAPDAILRNLTLPQPAYVPPARHRNEPFFSPKQTSSPQPDMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 90130..126591
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JMV71_RS24085 | 85386..86162 | + | 777 | WP_121572319 | hypothetical protein | - |
| JMV71_RS24090 | 86193..86462 | - | 270 | WP_048227650 | hypothetical protein | - |
| JMV71_RS24095 | 86535..88835 | - | 2301 | WP_053389033 | F-type conjugative transfer protein TrbC | - |
| JMV71_RS24100 | 88944..90074 | + | 1131 | WP_053389034 | RNA-guided endonuclease TnpB family protein | - |
| JMV71_RS24105 | 90130..91329 | - | 1200 | WP_053389035 | thioredoxin fold domain-containing protein | trbB |
| JMV71_RS24110 | 91382..92590 | - | 1209 | WP_053389036 | hypothetical protein | trbA |
| JMV71_RS24115 | 92893..93525 | - | 633 | WP_053069310 | plasmid IncI1-type surface exclusion protein ExcA | - |
| JMV71_RS24120 | 93587..95749 | - | 2163 | WP_053389037 | DotA/TraY family protein | traY |
| JMV71_RS24125 | 95806..96348 | - | 543 | WP_094935555 | conjugal transfer protein TraX | - |
| JMV71_RS24130 | 96345..97547 | - | 1203 | WP_048227644 | conjugal transfer protein TraW | traW |
| JMV71_RS24135 | 97560..98132 | - | 573 | WP_161556285 | conjugal transfer protein TraV | traV |
| JMV71_RS24140 | 98129..101182 | - | 3054 | WP_121572317 | ATP-binding protein | traU |
| JMV71_RS24145 | 101254..101985 | - | 732 | WP_069325142 | hypothetical protein | traT |
| JMV71_RS24150 | 102169..102573 | - | 405 | WP_048227642 | DUF6750 family protein | traR |
| JMV71_RS24155 | 102608..103132 | - | 525 | WP_048227641 | conjugal transfer protein TraQ | traQ |
| JMV71_RS24160 | 103132..103905 | - | 774 | WP_129719564 | conjugal transfer protein TraP | traP |
| JMV71_RS24165 | 103886..105202 | - | 1317 | WP_069325141 | conjugal transfer protein TraO | traO |
| JMV71_RS24170 | 105202..106185 | - | 984 | WP_048227638 | DotH/IcmK family type IV secretion protein | traN |
| JMV71_RS24175 | 106196..106897 | - | 702 | WP_048227637 | DotI/IcmL/TraM family protein | traM |
| JMV71_RS24180 | 106903..107253 | - | 351 | WP_048227636 | hypothetical protein | traL |
| JMV71_RS24185 | 108555..111770 | - | 3216 | Protein_119 | LPD7 domain-containing protein | - |
| JMV71_RS24190 | 111842..112129 | - | 288 | WP_048227635 | hypothetical protein | traK |
| JMV71_RS24195 | 112126..113286 | - | 1161 | WP_048227634 | plasmid transfer ATPase TraJ | virB11 |
| JMV71_RS24200 | 113298..114104 | - | 807 | WP_121572315 | type IV secretory system conjugative DNA transfer family protein | traI |
| JMV71_RS24205 | 114101..114574 | - | 474 | WP_121572314 | DotD/TraH family lipoprotein | - |
| JMV71_RS24210 | 114707..115915 | - | 1209 | WP_099530103 | conjugal transfer protein TraF | - |
| JMV71_RS24215 | 116018..116854 | - | 837 | WP_048227630 | hypothetical protein | traE |
| JMV71_RS24220 | 117000..117476 | - | 477 | WP_145996426 | hypothetical protein | - |
| JMV71_RS24225 | 117496..118656 | - | 1161 | WP_048227629 | site-specific integrase | - |
| JMV71_RS24230 | 119482..119865 | + | 384 | Protein_128 | phage tail protein | - |
| JMV71_RS24235 | 120247..120753 | - | 507 | WP_077223963 | sugar O-acetyltransferase | - |
| JMV71_RS24240 | 120808..122094 | - | 1287 | WP_058687259 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| JMV71_RS24245 | 122091..122765 | - | 675 | WP_121572313 | prepilin peptidase | - |
| JMV71_RS24250 | 122750..123232 | - | 483 | WP_069325157 | lytic transglycosylase domain-containing protein | virB1 |
| JMV71_RS24255 | 123317..123931 | - | 615 | WP_052463753 | type 4 pilus major pilin | - |
| JMV71_RS24260 | 123945..125039 | - | 1095 | WP_040231337 | type II secretion system F family protein | - |
| JMV71_RS24265 | 125032..126591 | - | 1560 | WP_048227627 | GspE/PulE family protein | virB11 |
| JMV71_RS24270 | 126594..127094 | - | 501 | WP_053069306 | type IV pilus biogenesis protein PilP | - |
| JMV71_RS24275 | 127078..128382 | - | 1305 | WP_048227626 | type 4b pilus protein PilO2 | - |
| JMV71_RS24280 | 128384..130060 | - | 1677 | WP_048227625 | PilN family type IVB pilus formation outer membrane protein | - |
| JMV71_RS24285 | 130075..130515 | - | 441 | WP_236582906 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 18932 | GenBank | NZ_LR890182 |
| Plasmid name | MSB1_1H|2 | Incompatibility group | - |
| Plasmid size | 135529 bp | Coordinate of oriT [Strand] | 80862..80941 [-] |
| Host baterium | Citrobacter freundii strain MSB1_1H isolate MSB1_1H |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |