Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118444 |
| Name | oriT_pRvVAR0630b |
| Organism | Agrobacterium vitis strain VAR06-30 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP023275 (274412..274448 [+], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 37 nt
GGCTCCAAGGGCGCAATTATACGTCGCTGATGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file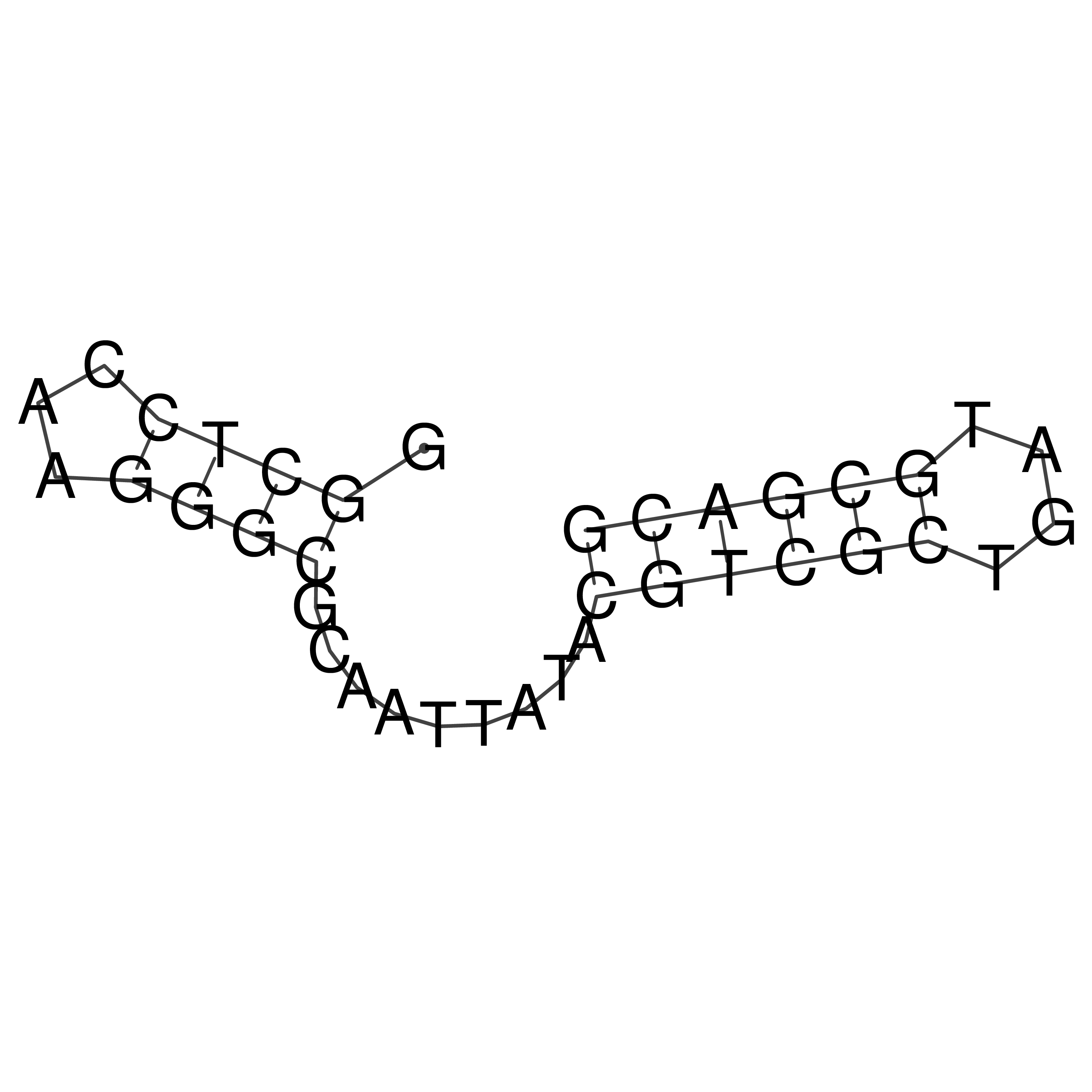
Relaxase
| ID | 11718 | GenBank | WP_180575622 |
| Name | traA_H1Y61_RS25365_pRvVAR0630b |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123490.61 Da Isoelectric Point: 9.6338
MAIAHFSASIISRGDGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHQEFVLPADAPNWVRSLIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTENGFGAKKVAVIGEDGQPVRTKSGKIVYELWAGSTEDFNKLRDGWFERQNHHLALSG
IDLRVDGRSYEKQGIDLEPTIHLGVGAKAIERKAESQGVRPELERIELNEKRRSENTRRILRRPEIVLDL
ITHEKSVFDEGDISKVLHRYIDDPGMFQQLMARIIQSPDVLRLKRDTIDFATGDKVPARYSTRAMIRLEA
RMATQAIWLSNREGRAVSPTILDETFRRHVRLSDEQRIAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWGRGRDMLNERTVFVMDEAGMVASKQMADFVD
AVVRTGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRHASLDFARGNVARALSAYQ
SEGKVLGSTLKAEAADHLISDWYRDYDPAKSTLILAHLRRDVGMLNGMARDKLVESGIIGEGHAFRTADG
IRQFDVGDQIVFLKNEGSLGVKNGMIGHVIEAAPNRIVAVVGEGDQRRQVTVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFAFNGGLTKVLSRRNAKETTLDYERGNLY
RGALRYAENRGLNIVQVARTLLRDQLDWTLRQKTRLADLAQRLRAIGERFGLQQSPKTQAMKETAPMVAG
IKTFSGSVADTVEQKLDSDPALKKQWEEVSTRFVYVYADPETAFRAMNFDAVLADKDSARQVLQTLEAEP
AFIGPLRGKTGILASKADREDRRVAELNVPVLKRDLEHYLRMRETAVQRLQADEQVLRQRISIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKTEIDGFNQALAQRFGERTLLTNAAREPSGKLFDTLSEG
MQPEQKERLKQAWPVMRAAQQLAAHERTVQSLKQSEELRLIQSQTSVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13720 | GenBank | WP_180575524 |
| Name | t4cp2_H1Y61_RS24830_pRvVAR0630b |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91776.74 Da Isoelectric Point: 5.5838
MVAPKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDGERNEVSRQINAILSRLG
SGWMIQVDAVRVPTGDYPDEAESHFPDPVTRAIDDERRAHFQKERGHFESRHALILTWRPPEPRRSGLAR
YVYSDAASRSATYADTMLQSFLTSIREIEQYLANVLSIRRMMTREISERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNTLDLMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDAIRLQEKCEAIRRLIQAEGFGARIEALNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQLFA
FDKGGSMLPLTLGLNGDHYQIGGDITPSAGAEVRMLAFCPLAELSTDGDRAWASEWIETLVALQGVTITP
DYRNAISRQIALMADSRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLSLGAFQCFEIEEL
MNMGERNLVPVLLYLFRRIEKRLTGAPSLIILDEAWLMLGHSVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNM
ALGPVALSFVGATGKEDLKRIRTLHSEHGAAWPCPWLQQRGIAHAEALFPTS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13721 | GenBank | WP_180575534 |
| Name | traG_H1Y61_RS25035_pRvVAR0630b |
UniProt ID | _ |
| Length | 640 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 640 a.a. Molecular weight: 68915.68 Da Isoelectric Point: 8.9882
MLAALVMTSGMEHRLAEFGSSTQAKLMLGRTGLALPYIAVAAIGAIALFAANGSADIKTVGVSVLAANSA
AIVIAMIRETIRLTGIASSVPAGQSVLTYADPATAVGAGVAFISAMFALRVTIKGNAAFAQTGPKRIGGK
RAVHGEADWMKLQEAAKLFPEPGGIVIGERYRVDKDSVATMPFRADEPQSWGAGGKSQLLCFDGSFGSSH
GIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDHRRKAGRKVIVLDPTLQGVGFNALDWIG
RHGNTKEEDIVAVATWIMTDNARTASVRDDFFRASAMQLLTALIADVCLSGHTDAKNQTLRRVRANLSEP
EPKLRGRLTEIYEQSESDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPHYAGLVSGDSFSTDDLAN
GGTDIFIALDLKVLEAHPGLARVVIGALLNAIYNRNGDVKGRTLFLLDEVARLGYLRILETARDAGRKYG
ITLTLIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTTVEVDQTNRSSGMKGS
SRSRSKQLSRRPLILPHELLRMRSDEQIVFTAGNAPLRCGRAIWFRREDMKACVGENRFHHPHTTTTAIT
APKVTKSDED
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13722 | GenBank | WP_180575579 |
| Name | traG_H1Y61_RS25350_pRvVAR0630b |
UniProt ID | _ |
| Length | 655 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 655 a.a. Molecular weight: 70542.70 Da Isoelectric Point: 9.1713
MTANRLLLFVLLAAIMLAALVMTSGMEHRLAEFGSSTQAKLMLGRTGLALPYIAVAAIGAIALFAANGSA
DIKTVGVSVLAANSAAIVIAMIRETIRLTGIASSVPAGQSVLTYADPATAVGAGVAFISAMFALRVTIKG
NAAFAQTGPKRIGGKRAVHGEADWMKLQEAAKLFPEPGGIVIGERYRVDKDSVATMPFRADEPQSWGAGG
KSQLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDHRRKAGRKVIVLD
PTLQGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNARTASVRDDFFRASAMQLLTALIADVCLSGHTDA
KNQTLRRVRANLSEPEPKLRGRLTEIYEQSESDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPHYA
GLVSGDSFSTDDLANGGTDIFIALDLKVLEAHPGLARVVIGALLNAIYNRNGDVKGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTLIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTAGNAPLRCGRAIWFRREDMKACVG
ENRFHHHHTTTTAIAASKVTKSDED
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 164653..182713
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H1Y61_RS24795 (RvVAR0630_pl05470) | 159972..161216 | - | 1245 | WP_180575517 | plasmid replication protein RepC | - |
| H1Y61_RS24800 (RvVAR0630_pl05480) | 161373..162368 | - | 996 | WP_180575518 | plasmid partitioning protein RepB | - |
| H1Y61_RS24805 (RvVAR0630_pl05490) | 162453..163643 | - | 1191 | WP_180575519 | plasmid partitioning protein RepA | - |
| H1Y61_RS24810 (RvVAR0630_pl05500) | 164018..164656 | + | 639 | WP_180575520 | acyl-homoserine-lactone synthase TraI | - |
| H1Y61_RS24815 (RvVAR0630_pl05510) | 164653..165624 | + | 972 | WP_180575521 | P-type conjugative transfer ATPase TrbB | virB11 |
| H1Y61_RS24820 (RvVAR0630_pl05520) | 165614..166018 | + | 405 | WP_180575522 | conjugal transfer pilin TrbC | virB2 |
| H1Y61_RS24825 (RvVAR0630_pl05530) | 166011..166310 | + | 300 | WP_180575523 | conjugal transfer protein TrbD | virB3 |
| H1Y61_RS24830 (RvVAR0630_pl05540) | 166321..168789 | + | 2469 | WP_180575524 | conjugal transfer protein TrbE | virb4 |
| H1Y61_RS24835 (RvVAR0630_pl05550) | 168761..169570 | + | 810 | WP_180575525 | P-type conjugative transfer protein TrbJ | virB5 |
| H1Y61_RS24840 | 169567..169794 | + | 228 | WP_180575526 | entry exclusion protein TrbK | - |
| H1Y61_RS24845 (RvVAR0630_pl05560) | 169788..170978 | + | 1191 | WP_180575527 | P-type conjugative transfer protein TrbL | virB6 |
| H1Y61_RS24850 (RvVAR0630_pl05570) | 170996..171658 | + | 663 | WP_180575528 | conjugal transfer protein TrbF | virB8 |
| H1Y61_RS24855 (RvVAR0630_pl05580) | 171736..172530 | + | 795 | WP_234644437 | P-type conjugative transfer protein TrbG | virB9 |
| H1Y61_RS24860 (RvVAR0630_pl05590) | 172530..173006 | + | 477 | WP_156556228 | conjugal transfer protein TrbH | - |
| H1Y61_RS24865 (RvVAR0630_pl05600) | 173018..174340 | + | 1323 | WP_180575529 | IncP-type conjugal transfer protein TrbI | virB10 |
| H1Y61_RS24870 | 174439..174735 | + | 297 | Protein_167 | DUF1403 family protein | - |
| H1Y61_RS24875 (RvVAR0630_pl05610) | 174851..175567 | - | 717 | WP_156619943 | IS6 family transposase | - |
| H1Y61_RS24880 (RvVAR0630_pl05620) | 175791..176294 | + | 504 | Protein_169 | OB-fold nucleic acid binding domain-containing protein | - |
| H1Y61_RS24885 (RvVAR0630_pl05630) | 176305..176970 | + | 666 | WP_180575530 | SOS response-associated peptidase family protein | - |
| H1Y61_RS24890 (RvVAR0630_pl05640) | 177000..177644 | + | 645 | WP_174113416 | SOS response-associated peptidase | - |
| H1Y61_RS24895 | 177652..177964 | - | 313 | Protein_172 | hypothetical protein | - |
| H1Y61_RS24900 (RvVAR0630_pl05670) | 177968..178204 | - | 237 | WP_235681004 | hypothetical protein | - |
| H1Y61_RS24905 (RvVAR0630_pl05680) | 178315..178485 | + | 171 | WP_156556177 | hypothetical protein | - |
| H1Y61_RS24910 (RvVAR0630_pl05690) | 178482..179288 | + | 807 | WP_174113417 | exodeoxyribonuclease III | - |
| H1Y61_RS24915 (RvVAR0630_pl05700) | 179278..179496 | + | 219 | WP_156556175 | hypothetical protein | - |
| H1Y61_RS24920 (RvVAR0630_pl05710) | 179657..180568 | - | 912 | WP_234644357 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| H1Y61_RS24925 (RvVAR0630_pl05720) | 180558..180956 | - | 399 | WP_234625163 | DUF6088 family protein | - |
| H1Y61_RS24930 (RvVAR0630_pl05730) | 181421..181600 | + | 180 | WP_180575531 | hypothetical protein | - |
| H1Y61_RS24935 (RvVAR0630_pl05740) | 181778..182713 | + | 936 | WP_156556169 | DUF3991 and toprim domain-containing protein | traP |
| H1Y61_RS24940 (RvVAR0630_pl05750) | 182846..183451 | + | 606 | WP_156556168 | DUF1419 domain-containing protein | - |
Host bacterium
| ID | 18877 | GenBank | NZ_AP023275 |
| Plasmid name | pRvVAR0630b | Incompatibility group | - |
| Plasmid size | 309504 bp | Coordinate of oriT [Strand] | 274412..274448 [+] |
| Host baterium | Agrobacterium vitis strain VAR06-30 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |