Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118411 |
| Name | oriT_AKSZ-142|unnamed1 |
| Organism | Enterococcus faecium strain AKSZ-142 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP110361 (2219..2422 [-], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
>oriT_AKSZ-142|unnamed1
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file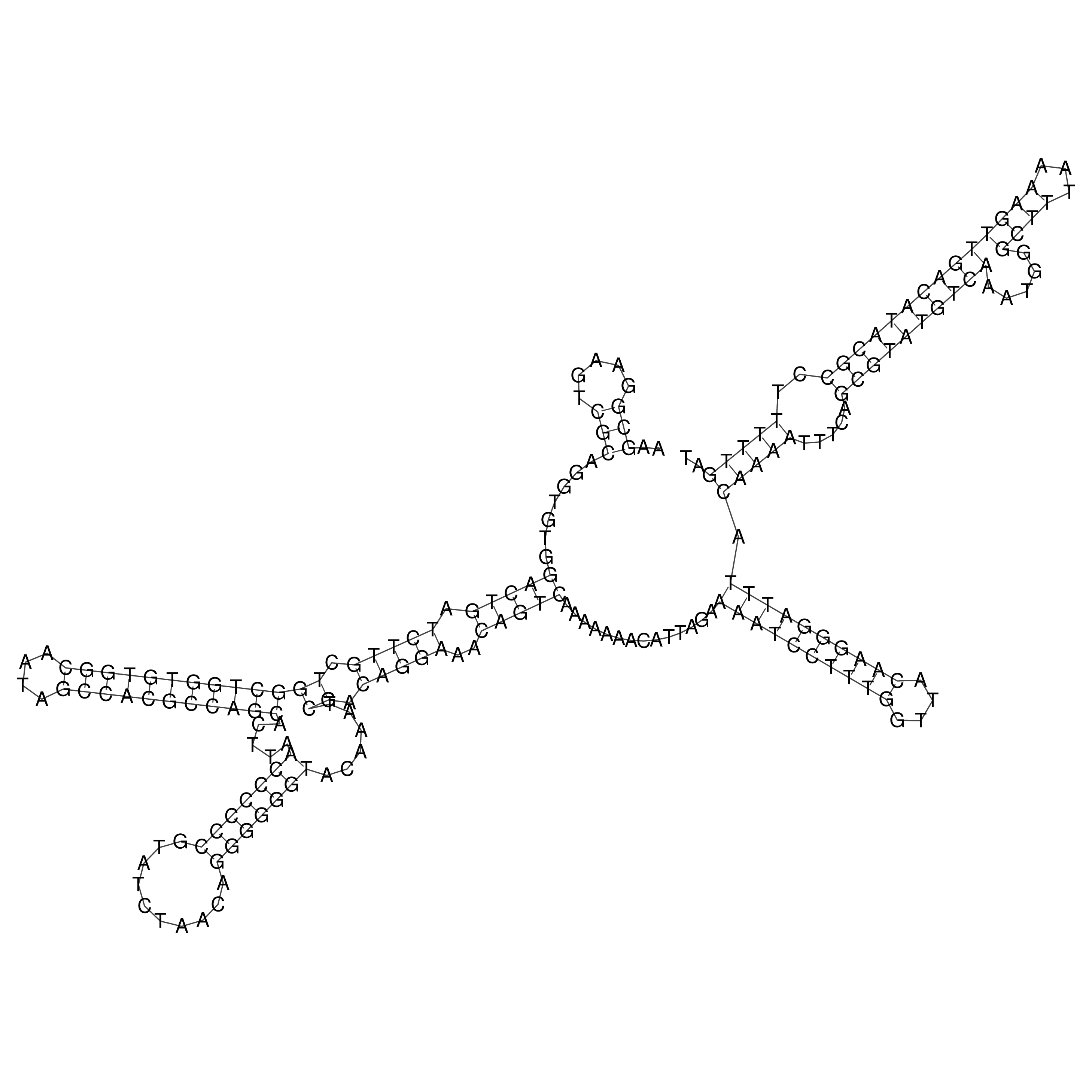
Relaxase
| ID | 11695 | GenBank | WP_000398284 |
| Name | mobT_MZO31_RS14425_AKSZ-142|unnamed1 |
UniProt ID | A0A3P3PY43 |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3PY43 |
T4CP
| ID | 13694 | GenBank | WP_000813488 |
| Name | tcpA_MZO31_RS14435_AKSZ-142|unnamed1 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>WP_000813488.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Bacteria]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 18844 | GenBank | NZ_CP110361 |
| Plasmid name | AKSZ-142|unnamed1 | Incompatibility group | - |
| Plasmid size | 54197 bp | Coordinate of oriT [Strand] | 2219..2422 [-] |
| Host baterium | Enterococcus faecium strain AKSZ-142 |
Cargo genes
| Drug resistance gene | erm(B), poxtA, fexB, tet(L), tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |