Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118337 |
| Name | oriT_pIMPIncN3_57kb |
| Organism | Enterobacter kobei strain EB_P8_L5_01.19 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP043516 (44216..44305 [+], 90 nt) |
| oriT length | 90 nt |
| IRs (inverted repeats) | 34..41, 44..51 (AGTGCGAT..ATCGCACT) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 90 nt
>oriT_pIMPIncN3_57kb
TATTTCATTTTTTGTTTTTTTAATCAGTTAGTTAGTGCGATTCATCGCACTGCGTTAGGTGTATAGCAGGTTAAGGGCTAAAAAATAATC
TATTTCATTTTTTGTTTTTTTAATCAGTTAGTTAGTGCGATTCATCGCACTGCGTTAGGTGTATAGCAGGTTAAGGGCTAAAAAATAATC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file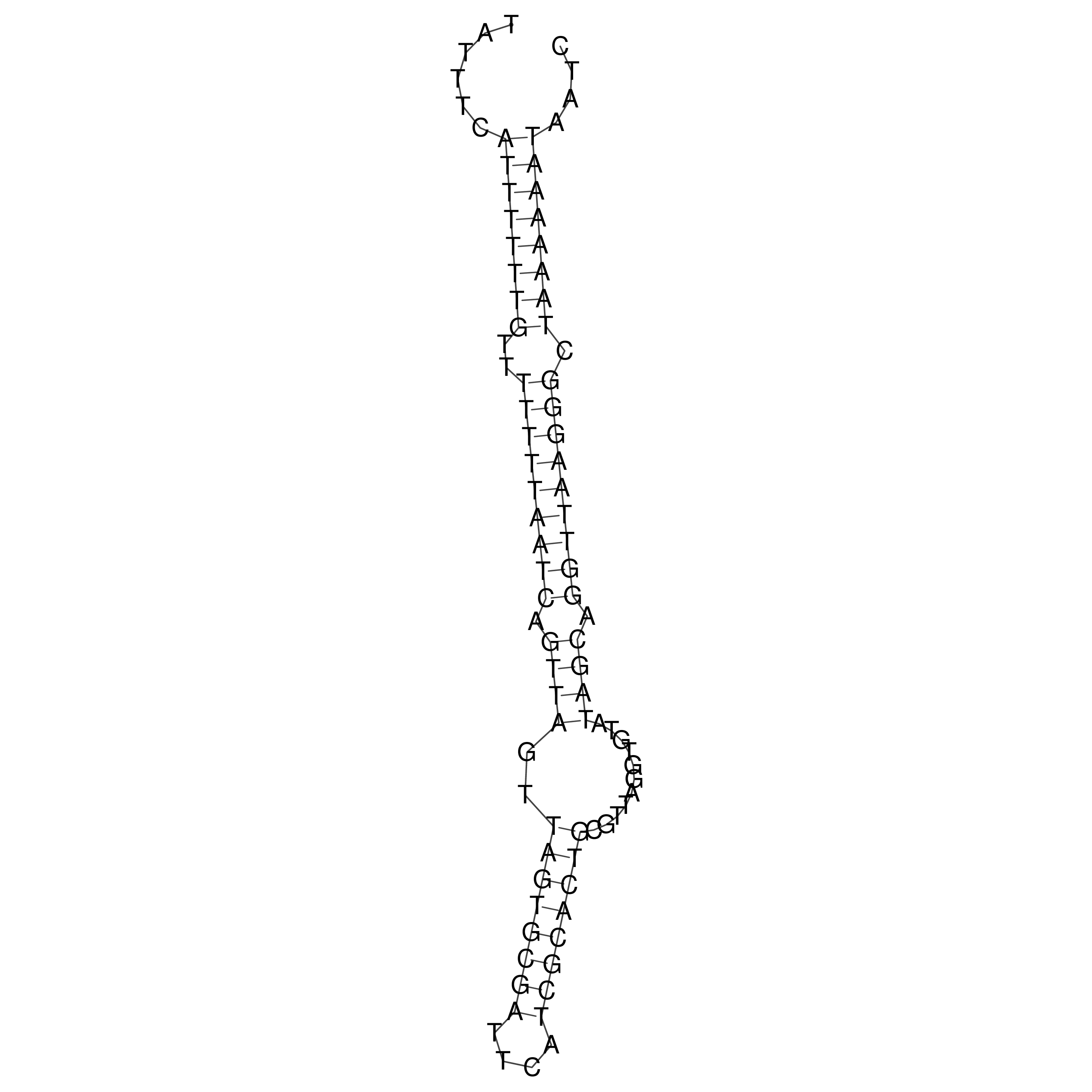
Relaxase
| ID | 11646 | GenBank | WP_238337819 |
| Name | mobF_FZO55_RS26490_pIMPIncN3_57kb |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 606 a.a. Molecular weight: 66540.30 Da Isoelectric Point: 9.6448
>WP_238337819.1 MobF family relaxase [Enterobacter kobei]
MLDITTITRIDIASVVGYYADAKDDYYSKDSSFTAWHGQGAEALGLSGEVSSQRFKELLVGEIDPFTQMK
RSSGDATKERLGYDLTFSAPKGVSMQALIHGDKAIIAAHEKAVAAAVQEAEKLAQARTTHAGKTITQNTG
NLVVASFRHETSRALDPELHTHAFVMNMTQRGDGQWRALKNDELMRAKMHLGDVYKQELALELTKAGYEL
RYNNKNNTFDMAHFTDEQIRGFSRRSAQIEEGLAAMGLTRETADAQTKSRVSMATRDKKTDDYSREDIHK
EWVNRANELGIDFNDLSWAGKGKDGPQTSMNTVPNFTSPEVKADRAIQFALKSLSERDASFDQSRLLAVA
NKKAIGQASKQDIEAAYYRAVAKGAIIEGEARYTTTTDQRSKDAPPLPAMTKAEWQAQLTDKKGMNNDQA
KALVAEGIKSGRLKKTSHRVTTVEGIRVEKAVLNIEARGRGKVIAALTSEQVTAQLVGKTLKAEQQKSVV
DICTTTDRFVAAHGFAGTGKSYMTMSAKAVLESQGFNVTALAPYGTQKKSLEDEGMPARTVAAFVKAKDK
KIDEKSVVFIDEAGVIPARQMKVLMETIEKAGARAVFWATPHRQKP
MLDITTITRIDIASVVGYYADAKDDYYSKDSSFTAWHGQGAEALGLSGEVSSQRFKELLVGEIDPFTQMK
RSSGDATKERLGYDLTFSAPKGVSMQALIHGDKAIIAAHEKAVAAAVQEAEKLAQARTTHAGKTITQNTG
NLVVASFRHETSRALDPELHTHAFVMNMTQRGDGQWRALKNDELMRAKMHLGDVYKQELALELTKAGYEL
RYNNKNNTFDMAHFTDEQIRGFSRRSAQIEEGLAAMGLTRETADAQTKSRVSMATRDKKTDDYSREDIHK
EWVNRANELGIDFNDLSWAGKGKDGPQTSMNTVPNFTSPEVKADRAIQFALKSLSERDASFDQSRLLAVA
NKKAIGQASKQDIEAAYYRAVAKGAIIEGEARYTTTTDQRSKDAPPLPAMTKAEWQAQLTDKKGMNNDQA
KALVAEGIKSGRLKKTSHRVTTVEGIRVEKAVLNIEARGRGKVIAALTSEQVTAQLVGKTLKAEQQKSVV
DICTTTDRFVAAHGFAGTGKSYMTMSAKAVLESQGFNVTALAPYGTQKKSLEDEGMPARTVAAFVKAKDK
KIDEKSVVFIDEAGVIPARQMKVLMETIEKAGARAVFWATPHRQKP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13632 | GenBank | WP_032493179 |
| Name | t4cp2_FZO55_RS25835_pIMPIncN3_57kb |
UniProt ID | _ |
| Length | 510 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 510 a.a. Molecular weight: 57912.96 Da Isoelectric Point: 9.5127
>WP_032493179.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MTEQERGLIFLCSLLGPIPFVWFITAKFVYGINSETAHYLVPYLLKNTFNLWPLWVAIIAGAVIGLCGFI
GFIIYDKGRVYKGPRFKRVLRGTKLVRASSLAEKTKERDHKQLTVAGVPVPTEYETLHFSIGGATGTGKS
TIIKEAIYSSLKRGTDKRIVLDPNGEYVETFYRPGDVILSPYDKRSPGWSMFNEIRDDFDYDLYAVSIIQ
ESKERSAEEWYRYGRLIYREVSKKLLTLKRSPTMEEVMHWCTIVENEKLKEFVKGTPAEALFIKGAERAT
GSARFVLGDKLAPHLKMPAGDFSLRDWLEDDKPGTLFITWQEQMKKSLNPLISCWLDIMFSSLLGMGKRD
QRTLFFIDELESLEYLPNLQDLLTKGRKSGASVWAGYQTYSQLIERYGINIAETLLGSMRSGIVMGSSRL
GKETLEQMSRALGEIEGEVEHKQGNPFQAGIGNRPRIETRTVRAVTPTEISELKNLTGYVYFPGDLPVGK
FKTKHVQYTRKNPVPGIIMR
MTEQERGLIFLCSLLGPIPFVWFITAKFVYGINSETAHYLVPYLLKNTFNLWPLWVAIIAGAVIGLCGFI
GFIIYDKGRVYKGPRFKRVLRGTKLVRASSLAEKTKERDHKQLTVAGVPVPTEYETLHFSIGGATGTGKS
TIIKEAIYSSLKRGTDKRIVLDPNGEYVETFYRPGDVILSPYDKRSPGWSMFNEIRDDFDYDLYAVSIIQ
ESKERSAEEWYRYGRLIYREVSKKLLTLKRSPTMEEVMHWCTIVENEKLKEFVKGTPAEALFIKGAERAT
GSARFVLGDKLAPHLKMPAGDFSLRDWLEDDKPGTLFITWQEQMKKSLNPLISCWLDIMFSSLLGMGKRD
QRTLFFIDELESLEYLPNLQDLLTKGRKSGASVWAGYQTYSQLIERYGINIAETLLGSMRSGIVMGSSRL
GKETLEQMSRALGEIEGEVEHKQGNPFQAGIGNRPRIETRTVRAVTPTEISELKNLTGYVYFPGDLPVGK
FKTKHVQYTRKNPVPGIIMR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4415..14691
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FZO55_RS25545 (FZO55_25545) | 45..1173 | - | 1129 | Protein_0 | IS3-like element ISSen4 family transposase | - |
| FZO55_RS25550 (FZO55_25550) | 1403..1735 | + | 333 | WP_032493174 | hypothetical protein | - |
| FZO55_RS25555 (FZO55_25555) | 1922..2530 | + | 609 | WP_000275218 | recombinase family protein | - |
| FZO55_RS25560 (FZO55_25560) | 2527..2712 | - | 186 | WP_024562121 | ribbon-helix-helix domain-containing protein | - |
| FZO55_RS25565 (FZO55_25565) | 2997..3305 | - | 309 | WP_000733764 | hypothetical protein | - |
| FZO55_RS25570 (FZO55_25570) | 3309..3647 | - | 339 | WP_032493175 | hypothetical protein | - |
| FZO55_RS25575 (FZO55_25575) | 3644..3994 | - | 351 | WP_024562120 | hypothetical protein | - |
| FZO55_RS25580 (FZO55_25580) | 3998..4315 | - | 318 | WP_000960955 | H-NS family nucleoid-associated regulatory protein | - |
| FZO55_RS25585 (FZO55_25585) | 4415..5197 | + | 783 | WP_024562119 | lytic transglycosylase domain-containing protein | virB1 |
| FZO55_RS25595 (FZO55_25595) | 5444..5740 | + | 297 | WP_015060075 | TrbC/VirB2 family protein | virB2 |
| FZO55_RS25600 (FZO55_25600) | 5780..6097 | + | 318 | WP_000462629 | type IV secretion system protein VirB3 | virB3 |
| FZO55_RS25605 (FZO55_25605) | 6098..8671 | + | 2574 | WP_040115085 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| FZO55_RS25610 (FZO55_25610) | 8687..9388 | + | 702 | WP_014014949 | type IV secretion system protein | virB5 |
| FZO55_RS25615 (FZO55_25615) | 9398..9640 | + | 243 | WP_000858958 | EexN family lipoprotein | - |
| FZO55_RS25620 (FZO55_25620) | 9657..10694 | + | 1038 | WP_014014948 | type IV secretion system protein | virB6 |
| FZO55_RS25630 (FZO55_25630) | 10913..11608 | + | 696 | WP_001208352 | type IV secretion system protein | virB8 |
| FZO55_RS25635 (FZO55_25635) | 11619..12506 | + | 888 | WP_000758230 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| FZO55_RS25640 (FZO55_25640) | 12506..13639 | + | 1134 | WP_000101920 | type IV secretion system protein VirB10 | virB10 |
| FZO55_RS25645 (FZO55_25645) | 13690..14691 | + | 1002 | WP_001076634 | ATPase, T2SS/T4P/T4SS family | virB11 |
| FZO55_RS25650 (FZO55_25650) | 14703..15215 | + | 513 | WP_029697418 | phospholipase D family protein | - |
| FZO55_RS25655 (FZO55_25655) | 15724..16728 | - | 1005 | WP_000427620 | IS110-like element IS4321 family transposase | - |
Host bacterium
| ID | 18770 | GenBank | NZ_CP043516 |
| Plasmid name | pIMPIncN3_57kb | Incompatibility group | IncN3 |
| Plasmid size | 56515 bp | Coordinate of oriT [Strand] | 44216..44305 [+] |
| Host baterium | Enterobacter kobei strain EB_P8_L5_01.19 |
Cargo genes
| Drug resistance gene | blaIMP-70, qacE, sul1, qnrA1 |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merC, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |