Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118331 |
| Name | oriT_pAvCG78b |
| Organism | Agrobacterium vitis strain CG78 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP056050 (284430..284466 [-], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 37 nt
>oriT_pAvCG78b
GGCTCCAAGGGCGCAATTATACGTCGCTGATGCGACG
GGCTCCAAGGGCGCAATTATACGTCGCTGATGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file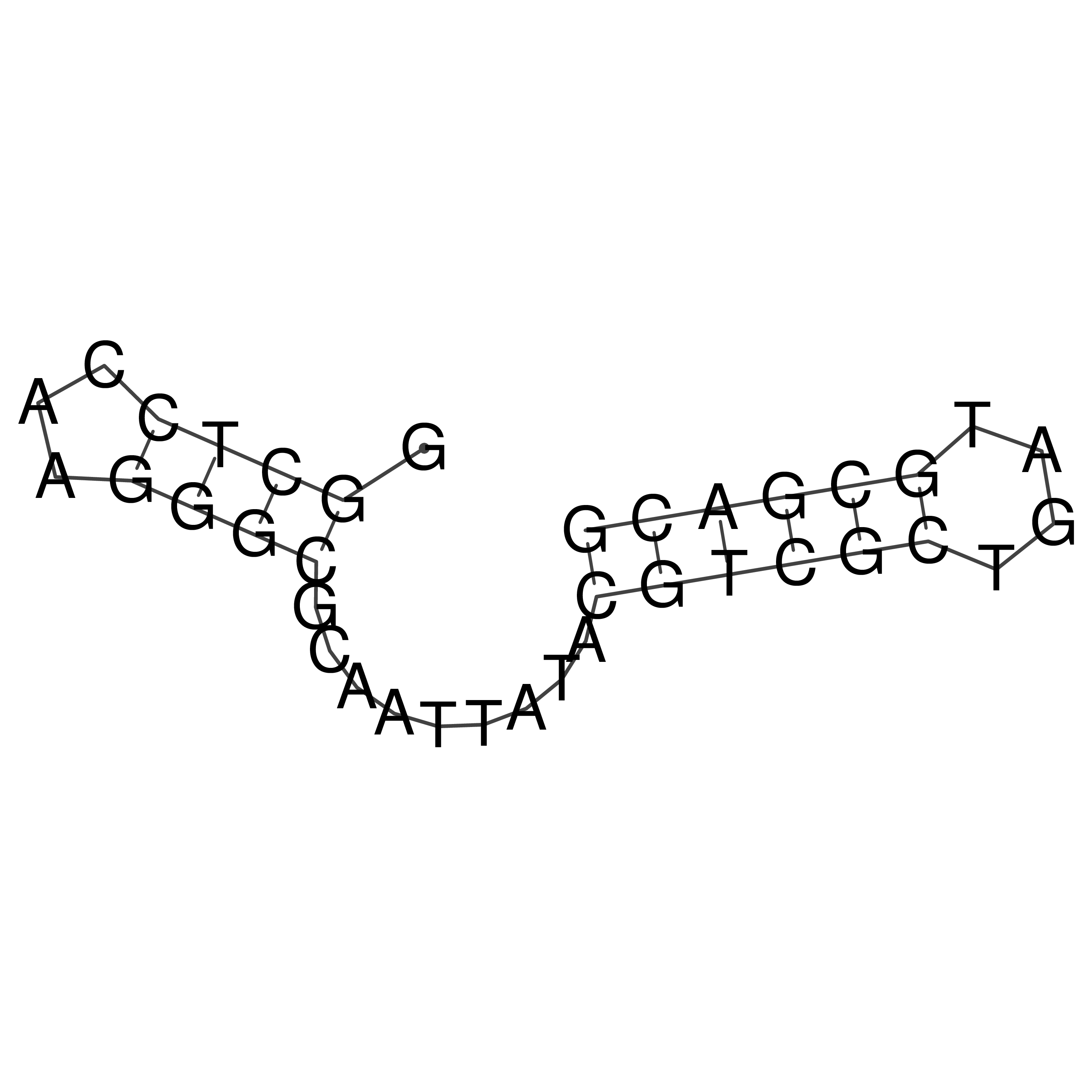
Relaxase
| ID | 11641 | GenBank | WP_071205421 |
| Name | traA_AVCG78_RS25525_pAvCG78b |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123909.64 Da Isoelectric Point: 9.9753
>WP_071205421.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium/Agrobacterium group]
MAIAHFSASIISRGDGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHQEFVLPADAPNWVRSLIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLSEDGFGAKKVAVIGEDGQPVRTKSGKIVYELWAGSTEDFNALRDGWFERLNHHLALRG
FDLRVDGRSYEKQGIDLEPTIHLGVGTKAIERKAKSKALRPELERIELNEQLRRDNARRILRKPGIVLDM
ITRERSVFDERDVAKVLHRYVDDPAVFQQLMLRIIQNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMVRQALWLSNRYGHDVSDAALNATFRRHERLSDEQKTAIELIARPSRIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDRTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRALVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFKTADG
VRQFDVGDQIVFLKNETSLGVKNGMIAQVIEAAPNRIVAIIGEGDERRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAESRGLNIVQVARTLVRDQLDWTLRQKTKLVDLGQRLASFAERLGLHHPPKTQTIKEAAPMVAG
IKTFSGSVADTVEHKLDTDPALRQQWEEVSTRFSYVFADPETAFRAMNFDAVLADKETAKLALQTLTTAP
VSIGPLRGKTGILASKADREHRRVAELNVPALKRDLEHYLRMRETAVQRLKADEQVLRQRISIDIPALSP
AAHVVLERVRYAIDRNDLPAAMANALSNRETKLEIDGFNQAVTQRFGERTLLTNAAREPSGKIFDTLSNG
MEPEQKERLKQAWPVMRAAQQLAAHERTVQSLKQSEELRLIQSQTSVLKQ
MAIAHFSASIISRGDGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHQEFVLPADAPNWVRSLIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLSEDGFGAKKVAVIGEDGQPVRTKSGKIVYELWAGSTEDFNALRDGWFERLNHHLALRG
FDLRVDGRSYEKQGIDLEPTIHLGVGTKAIERKAKSKALRPELERIELNEQLRRDNARRILRKPGIVLDM
ITRERSVFDERDVAKVLHRYVDDPAVFQQLMLRIIQNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMVRQALWLSNRYGHDVSDAALNATFRRHERLSDEQKTAIELIARPSRIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDDRTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRALVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFKTADG
VRQFDVGDQIVFLKNETSLGVKNGMIAQVIEAAPNRIVAIIGEGDERRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAESRGLNIVQVARTLVRDQLDWTLRQKTKLVDLGQRLASFAERLGLHHPPKTQTIKEAAPMVAG
IKTFSGSVADTVEHKLDTDPALRQQWEEVSTRFSYVFADPETAFRAMNFDAVLADKETAKLALQTLTTAP
VSIGPLRGKTGILASKADREHRRVAELNVPALKRDLEHYLRMRETAVQRLKADEQVLRQRISIDIPALSP
AAHVVLERVRYAIDRNDLPAAMANALSNRETKLEIDGFNQAVTQRFGERTLLTNAAREPSGKIFDTLSNG
MEPEQKERLKQAWPVMRAAQQLAAHERTVQSLKQSEELRLIQSQTSVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13622 | GenBank | WP_071585024 |
| Name | traG_AVCG78_RS25540_pAvCG78b |
UniProt ID | _ |
| Length | 655 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 655 a.a. Molecular weight: 70556.77 Da Isoelectric Point: 9.1709
>WP_071585024.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium/Agrobacterium group]
MTANRLLLFVLLAAIMLAALVMTSGMEHRLAEFGSSTQAKLMLGRTGLALPYIAVAAIGAIALFAANGSA
DIKTVGVSVLAANSAAIVIAMIRETIRLTGIASSVPAGQSVLTYADPATAVGAGVAFISAMFALRVTIKG
NAAFAQTGPKRIGGKRAVHGEADWMKLQEAAKLFPEPGGIVIGERYRVDKDSVATMPFRADEPQSWGAGG
KSQLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDHRRKAGRKVIVLD
PTLQGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNARTASVRDDFFRASAMQLLTALIADVCLSGHTDA
KNQTLRRVRANLSEPEPKLRGRLTEIYEQSESDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPHYA
GLVSGDSFSTDDLANGGTDIFIALDLKVLEAHPGLARVVIGALLNAIYNRNGDVKGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTLIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLSRRPLILPHELLRMRSDEQIVFTAGNAPLRCGRAIWFRREDMKACVG
ENRFHHPHTTTTAITAPKVTKSDED
MTANRLLLFVLLAAIMLAALVMTSGMEHRLAEFGSSTQAKLMLGRTGLALPYIAVAAIGAIALFAANGSA
DIKTVGVSVLAANSAAIVIAMIRETIRLTGIASSVPAGQSVLTYADPATAVGAGVAFISAMFALRVTIKG
NAAFAQTGPKRIGGKRAVHGEADWMKLQEAAKLFPEPGGIVIGERYRVDKDSVATMPFRADEPQSWGAGG
KSQLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDHRRKAGRKVIVLD
PTLQGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNARTASVRDDFFRASAMQLLTALIADVCLSGHTDA
KNQTLRRVRANLSEPEPKLRGRLTEIYEQSESDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPHYA
GLVSGDSFSTDDLANGGTDIFIALDLKVLEAHPGLARVVIGALLNAIYNRNGDVKGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTLIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLSRRPLILPHELLRMRSDEQIVFTAGNAPLRCGRAIWFRREDMKACVG
ENRFHHPHTTTTAITAPKVTKSDED
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13623 | GenBank | WP_071204871 |
| Name | t4cp2_AVCG78_RS25925_pAvCG78b |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91967.89 Da Isoelectric Point: 5.8031
>WP_071204871.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium/Agrobacterium group]
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAIRVPTGDYPTEAESHFSDPVTRAIDDERRAHFQKERGHFESRHALILTWRPPEPRRSGLTR
YIYSDAASRSATYADKALESFLTSIREVEQYLANVVSIRRMMTRETPERGGFRIARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDATRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPVAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTLGLDGDHYQIGGDIGASTVEEGKVLAFCPLAELSTDGDRAWASEWIEMLVALQGVTITP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLSLGAFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNM
ALGPVALSFVGATGKEDLKRIRSSHSQHGAAWPLHWLQQRGIAYAEKLFSAE
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAIRVPTGDYPTEAESHFSDPVTRAIDDERRAHFQKERGHFESRHALILTWRPPEPRRSGLTR
YIYSDAASRSATYADKALESFLTSIREVEQYLANVVSIRRMMTRETPERGGFRIARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDATRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPVAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTLGLDGDHYQIGGDIGASTVEEGKVLAFCPLAELSTDGDRAWASEWIEMLVALQGVTITP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLSLGAFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNM
ALGPVALSFVGATGKEDLKRIRSSHSQHGAAWPLHWLQQRGIAYAEKLFSAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 360114..369812
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AVCG78_RS25865 (AVCG78_25840) | 356192..356686 | + | 495 | WP_071205479 | hypothetical protein | - |
| AVCG78_RS25870 (AVCG78_25845) | 356799..356957 | + | 159 | WP_156582886 | hypothetical protein | - |
| AVCG78_RS25875 (AVCG78_25850) | 357243..358064 | + | 822 | WP_071205480 | LON peptidase substrate-binding domain-containing protein | - |
| AVCG78_RS25880 (AVCG78_25855) | 358361..359059 | - | 699 | WP_071205481 | SMC-Scp complex subunit ScpB | - |
| AVCG78_RS25885 (AVCG78_25860) | 359062..360015 | - | 954 | WP_071205482 | DUF1403 family protein | - |
| AVCG78_RS25890 (AVCG78_25865) | 360114..361436 | - | 1323 | WP_071204877 | IncP-type conjugal transfer protein TrbI | virB10 |
| AVCG78_RS25895 (AVCG78_25870) | 361448..361924 | - | 477 | WP_071204876 | conjugal transfer protein TrbH | - |
| AVCG78_RS25900 (AVCG78_25875) | 361924..362718 | - | 795 | WP_234618193 | P-type conjugative transfer protein TrbG | virB9 |
| AVCG78_RS25905 (AVCG78_25880) | 362793..363470 | - | 678 | WP_071204875 | conjugal transfer protein TrbF | virB8 |
| AVCG78_RS25910 (AVCG78_25885) | 363488..364678 | - | 1191 | WP_071585038 | P-type conjugative transfer protein TrbL | virB6 |
| AVCG78_RS25915 (AVCG78_25890) | 364672..364899 | - | 228 | WP_071204873 | entry exclusion protein TrbK | - |
| AVCG78_RS25920 (AVCG78_25895) | 364896..365663 | - | 768 | WP_234618192 | P-type conjugative transfer protein TrbJ | virB5 |
| AVCG78_RS25925 (AVCG78_25900) | 365677..368145 | - | 2469 | WP_071204871 | conjugal transfer protein TrbE | virb4 |
| AVCG78_RS25930 (AVCG78_25905) | 368167..368454 | - | 288 | WP_071204870 | conjugal transfer protein TrbD | virB3 |
| AVCG78_RS25935 (AVCG78_25910) | 368447..368851 | - | 405 | WP_071204869 | conjugal transfer pilin TrbC | virB2 |
| AVCG78_RS25940 (AVCG78_25915) | 368841..369812 | - | 972 | WP_071204868 | P-type conjugative transfer ATPase TrbB | virB11 |
| AVCG78_RS25945 (AVCG78_25920) | 369809..370447 | - | 639 | WP_071585036 | acyl-homoserine-lactone synthase TraI | - |
Host bacterium
| ID | 18764 | GenBank | NZ_CP056050 |
| Plasmid name | pAvCG78b | Incompatibility group | - |
| Plasmid size | 370903 bp | Coordinate of oriT [Strand] | 284430..284466 [-] |
| Host baterium | Agrobacterium vitis strain CG78 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bphC |
| Symbiosis gene | - |
| Anti-CRISPR | - |