Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118296 |
| Name | oriT_UP_1435|unnamed1 |
| Organism | Staphylococcus aureus strain UP_1435 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP047857 (24746..24934 [+], 189 nt) |
| oriT length | 189 nt |
| IRs (inverted repeats) | 149..155, 162..168 (CTATCAT..ATGATAG) 132..138, 142..148 (GTCTGGC..GCCAGAC) 62..70, 77..85 (TAGTGTCAC..GTGACACTA) 33..38, 45..50 (TGTCAC..GTGACA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 189 nt
>oriT_UP_1435|unnamed1
TGTCACAAAAATAAGACAAAATCTACATGTAGTGTCACAAAACCGTGACAAACACAATATATAGTGTCACGAAAATGTGACACTAGGTGTTTTTATGATATCACTATGAAATGTCCTAAAACCCTTGGAATGTCTGGCTTTGCCAGACCTATCATTGTCCGATGATAGCAAATTCCCCTTATGCTCTTA
TGTCACAAAAATAAGACAAAATCTACATGTAGTGTCACAAAACCGTGACAAACACAATATATAGTGTCACGAAAATGTGACACTAGGTGTTTTTATGATATCACTATGAAATGTCCTAAAACCCTTGGAATGTCTGGCTTTGCCAGACCTATCATTGTCCGATGATAGCAAATTCCCCTTATGCTCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file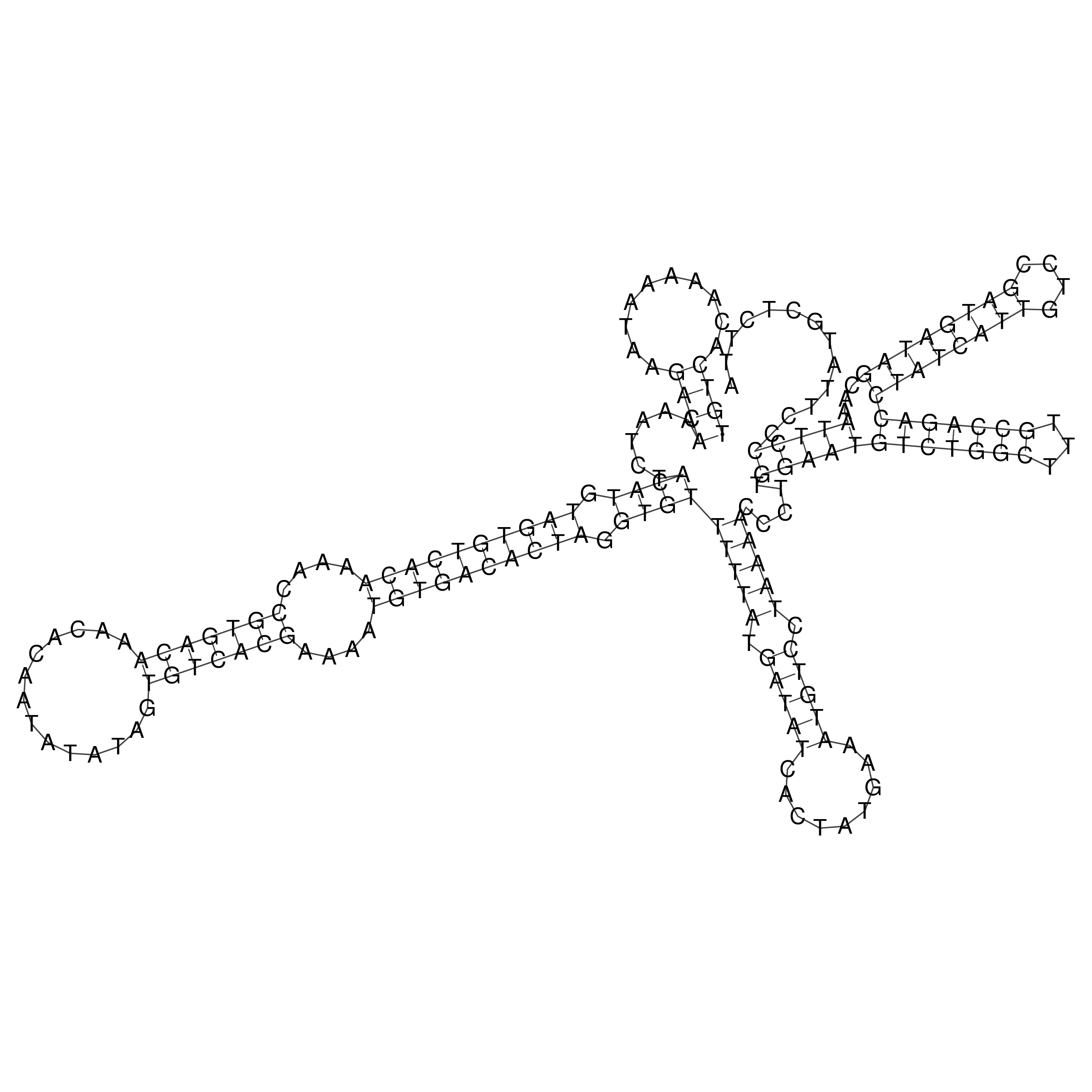
Relaxase
| ID | 11623 | GenBank | WP_160176696 |
| Name | mobP2_E3S66_RS14535_UP_1435|unnamed1 |
UniProt ID | _ |
| Length | 382 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 382 a.a. Molecular weight: 45664.89 Da Isoelectric Point: 9.9507
>WP_160176696.1 MobP2 family relaxase [Staphylococcus aureus]
MPKIVTVSQFEQPNEFYEYSNYVNYMKRGEAQNNKDDFKYDVYAHYMFDEEKSQNMFNEHDNFMSKKAIN
KIKSDFSHAQKNKGMMWKDVISFDNSSLESVGIYDSKTHTLDEQKIKEATRKMMKRFEKKEGLEDNLVWS
GAIHYNTDNIHVHVAAVEKVIKRTRGKRKPATLKHMKSTFANELFDIKGERQKINEFIRERIVKGIRDES
EPCKDKAMKKQIKKVHKLLEDIPRNQRNYNNNILKYIRPEIDKITDIYIKNYHSKDFEVFKNDLKRQTNL
YKETYGENSNYGSYEKTKMKDLYARSGNTILKNIKELDQDLMKVKQQSTHSKSNATITKLKIGKCLNQAL
YRTKYALRSDYDKEKNIQQYYQEFDKQSYRER
MPKIVTVSQFEQPNEFYEYSNYVNYMKRGEAQNNKDDFKYDVYAHYMFDEEKSQNMFNEHDNFMSKKAIN
KIKSDFSHAQKNKGMMWKDVISFDNSSLESVGIYDSKTHTLDEQKIKEATRKMMKRFEKKEGLEDNLVWS
GAIHYNTDNIHVHVAAVEKVIKRTRGKRKPATLKHMKSTFANELFDIKGERQKINEFIRERIVKGIRDES
EPCKDKAMKKQIKKVHKLLEDIPRNQRNYNNNILKYIRPEIDKITDIYIKNYHSKDFEVFKNDLKRQTNL
YKETYGENSNYGSYEKTKMKDLYARSGNTILKNIKELDQDLMKVKQQSTHSKSNATITKLKIGKCLNQAL
YRTKYALRSDYDKEKNIQQYYQEFDKQSYRER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13607 | GenBank | WP_001210915 |
| Name | t4cp2_E3S66_RS14485_UP_1435|unnamed1 |
UniProt ID | _ |
| Length | 923 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 923 a.a. Molecular weight: 106091.07 Da Isoelectric Point: 8.0720
>WP_001210915.1 VirD4-like conjugal transfer protein, CD1115 family [Staphylococcus aureus]
MRFGSKDKHGYRRKKLSFSQNKHLLAHPCILVSTLLLIMLLATAIANFLINILMTVFFGNHDIFTNMKQL
NTKYTDYIFNFRSDAVLLYIPIWGLVFIFFGKKVYQFRQRFKSLNNNEKASGRFATRKEITQQYKAIPQR
EHAFEGEGGLPVAMFGVYDFVTKTKQYLEQRKTAIREINGITDLTADEKLELRKDKLGKMNKEFATTFFS
KREFTFIDESPTNNLIVGTSRSGKGEIVVLSMIENYSRSSEQPSLIVNDMKGELFSASYKTLENRGYLVE
IFNLDQPMESTMSFNPLQQVIDEYMKGDLGEAQEMTKQITYTLTNNEGVEDNEWNLMAGALINAMILALC
EETLPKNPEKVTLYSVSNMLQTLSTKRFYKEIAIGNKVQLIEVSALDEYMERFPSHSPAKTQYATVQAAP
DKMRSSIIGTALKALQPFTTDTIAKLTSHTSFDINKLGFPSIIDGYATKDSTFELGVQVLRDGENGIEYE
EVEGIQIGVNEYGYWQYPFKTVLKVGDRIETIEKDGNNQVITSYFYVDHIDDKGQVTFNQKGELDNSDIT
IRKFNSFPKPIAVFMVVPDYNNANHGIASILVNQIVFEISKNSQLYTENQSTYRRVIFHLDEAGNMPPIP
NLSQKVNVSLSRGLRFNFFVQAFSQFKDKYGDAYDAIMDACQNKVYIMATQEQTLKDFSAMIGSQQITVH
SRSGEAGAIKSSITENQEERPLLRPDELARLQEGETVVVRNLKRQDKKRNKVMSYPIFNDGKYAMKYRYQ
YLSDLMDTSQSIIHFRPMLKARCEHRHLQLEATLVDWDKRIEEMQEGIDGQKEENKNINDINQYKKSNEE
GNVAVRVKRLETLKDKVGDAVFDKITRRFERYLRTYAEQGKNVNTITQKKLEELVLADSEVKEEEKTKRI
EMIKKLIKEAKTT
MRFGSKDKHGYRRKKLSFSQNKHLLAHPCILVSTLLLIMLLATAIANFLINILMTVFFGNHDIFTNMKQL
NTKYTDYIFNFRSDAVLLYIPIWGLVFIFFGKKVYQFRQRFKSLNNNEKASGRFATRKEITQQYKAIPQR
EHAFEGEGGLPVAMFGVYDFVTKTKQYLEQRKTAIREINGITDLTADEKLELRKDKLGKMNKEFATTFFS
KREFTFIDESPTNNLIVGTSRSGKGEIVVLSMIENYSRSSEQPSLIVNDMKGELFSASYKTLENRGYLVE
IFNLDQPMESTMSFNPLQQVIDEYMKGDLGEAQEMTKQITYTLTNNEGVEDNEWNLMAGALINAMILALC
EETLPKNPEKVTLYSVSNMLQTLSTKRFYKEIAIGNKVQLIEVSALDEYMERFPSHSPAKTQYATVQAAP
DKMRSSIIGTALKALQPFTTDTIAKLTSHTSFDINKLGFPSIIDGYATKDSTFELGVQVLRDGENGIEYE
EVEGIQIGVNEYGYWQYPFKTVLKVGDRIETIEKDGNNQVITSYFYVDHIDDKGQVTFNQKGELDNSDIT
IRKFNSFPKPIAVFMVVPDYNNANHGIASILVNQIVFEISKNSQLYTENQSTYRRVIFHLDEAGNMPPIP
NLSQKVNVSLSRGLRFNFFVQAFSQFKDKYGDAYDAIMDACQNKVYIMATQEQTLKDFSAMIGSQQITVH
SRSGEAGAIKSSITENQEERPLLRPDELARLQEGETVVVRNLKRQDKKRNKVMSYPIFNDGKYAMKYRYQ
YLSDLMDTSQSIIHFRPMLKARCEHRHLQLEATLVDWDKRIEEMQEGIDGQKEENKNINDINQYKKSNEE
GNVAVRVKRLETLKDKVGDAVFDKITRRFERYLRTYAEQGKNVNTITQKKLEELVLADSEVKEEEKTKRI
EMIKKLIKEAKTT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 18729 | GenBank | NZ_CP047857 |
| Plasmid name | UP_1435|unnamed1 | Incompatibility group | - |
| Plasmid size | 34909 bp | Coordinate of oriT [Strand] | 24746..24934 [+] |
| Host baterium | Staphylococcus aureus strain UP_1435 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |