Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118264 |
| Name | oriT_pSTN0717-53-3 |
| Organism | Enterobacter kobei strain STN0717-53 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP022501 (60805..60861 [-], 57 nt) |
| oriT length | 57 nt |
| IRs (inverted repeats) | 13..20, 23..30 (GCAAAATT..AATTTTGC) |
| Location of nic site | 39..40 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 57 nt
>oriT_pSTN0717-53-3
TAAAGTTAATCAGCAAAATTTCAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
TAAAGTTAATCAGCAAAATTTCAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file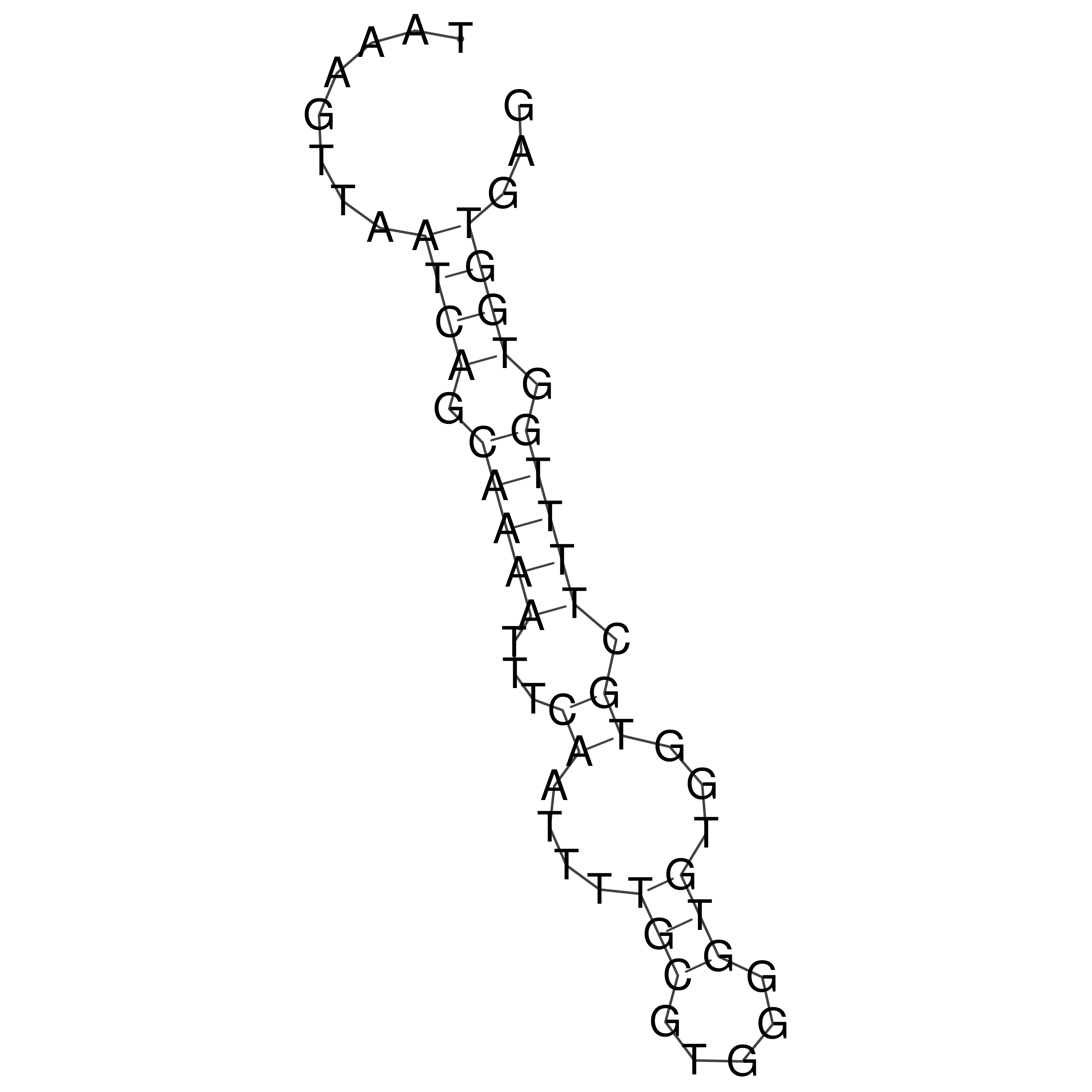
Relaxase
| ID | 11609 | GenBank | WP_200925823 |
| Name | traI_JLH17_RS26280_pSTN0717-53-3 |
UniProt ID | _ |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 188936.04 Da Isoelectric Point: 6.1461
>WP_200925823.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacter cloacae complex]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKATGFDIQAYREAADQRVVQGNIPATTPEAIDINSSVGQSIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRAGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGATGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGITFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVSGPREQAVLAGAVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVQLYSAQTAQKTTEKLSRQSAWSVVTSQVKE
AAGKDALGDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQGSLSLQAIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIERVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQIRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVSAGRAALRQAGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQVSRNGESLLARDMEEGVRLARENPQSGVVVRLEGEVRPWN
PGAITGGRVWADGLPDGTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMERIKETPASPLTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKATGFDIQAYREAADQRVVQGNIPATTPEAIDINSSVGQSIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRAGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGATGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGITFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVSGPREQAVLAGAVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVQLYSAQTAQKTTEKLSRQSAWSVVTSQVKE
AAGKDALGDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQGSLSLQAIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIERVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQIRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVSAGRAALRQAGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQVSRNGESLLARDMEEGVRLARENPQSGVVVRLEGEVRPWN
PGAITGGRVWADGLPDGTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMERIKETPASPLTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13594 | GenBank | WP_200928253 |
| Name | traC_JLH17_RS26200_pSTN0717-53-3 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98854.55 Da Isoelectric Point: 5.7676
>WP_200928253.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacter]
MSNPLDTVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYAGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAR
YTQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDTVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYAGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAR
YTQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13595 | GenBank | WP_200928256 |
| Name | traD_JLH17_RS26275_pSTN0717-53-3 |
UniProt ID | _ |
| Length | 742 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 742 a.a. Molecular weight: 83316.94 Da Isoelectric Point: 5.0983
>WP_200928256.1 type IV conjugative transfer system coupling protein TraD [Enterobacter kobei]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEGSAENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGIDESGEVVDM
AAWEAWQAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEGSAENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGIDESGEVVDM
AAWEAWQAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 60257..85984
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JLH17_RS26090 (STN0717ENT53_P30660) | 55355..55678 | + | 324 | WP_008324183 | hypothetical protein | - |
| JLH17_RS26095 (STN0717ENT53_P30670) | 55740..56099 | + | 360 | WP_008324181 | hypothetical protein | - |
| JLH17_RS26100 (STN0717ENT53_P30680) | 56726..57100 | + | 375 | WP_008324180 | hypothetical protein | - |
| JLH17_RS26105 (STN0717ENT53_P30690) | 57154..57441 | + | 288 | WP_008324178 | hypothetical protein | - |
| JLH17_RS26110 (STN0717ENT53_P30700) | 57488..57832 | + | 345 | WP_008324177 | hypothetical protein | - |
| JLH17_RS26115 (STN0717ENT53_P30710) | 57918..58181 | + | 264 | WP_008324174 | hypothetical protein | - |
| JLH17_RS26120 (STN0717ENT53_P30720) | 58224..59053 | + | 830 | Protein_75 | N-6 DNA methylase | - |
| JLH17_RS26125 (STN0717ENT53_P30730) | 59680..60210 | + | 531 | WP_008324170 | antirestriction protein | - |
| JLH17_RS26130 (STN0717ENT53_P30740) | 60257..60733 | - | 477 | WP_200928251 | transglycosylase SLT domain-containing protein | virB1 |
| JLH17_RS26135 (STN0717ENT53_P30750) | 61165..61560 | + | 396 | WP_125885549 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| JLH17_RS26140 (STN0717ENT53_P30760) | 61745..62449 | + | 705 | WP_045354209 | hypothetical protein | - |
| JLH17_RS26145 (STN0717ENT53_P30770) | 62558..62932 | + | 375 | WP_040218544 | TraY domain-containing protein | - |
| JLH17_RS26150 (STN0717ENT53_P30780) | 62998..63348 | + | 351 | WP_182930703 | type IV conjugative transfer system pilin TraA | - |
| JLH17_RS26155 (STN0717ENT53_P30790) | 63499..63804 | + | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| JLH17_RS26160 (STN0717ENT53_P30800) | 63819..64385 | + | 567 | WP_008324158 | type IV conjugative transfer system protein TraE | traE |
| JLH17_RS26165 (STN0717ENT53_P30810) | 64372..65115 | + | 744 | WP_011201807 | type-F conjugative transfer system secretin TraK | traK |
| JLH17_RS26170 (STN0717ENT53_P30820) | 65102..66496 | + | 1395 | WP_200928252 | F-type conjugal transfer pilus assembly protein TraB | traB |
| JLH17_RS26175 (STN0717ENT53_P30830) | 66518..67057 | + | 540 | WP_060446828 | type IV conjugative transfer system lipoprotein TraV | traV |
| JLH17_RS26180 (STN0717ENT53_P30840) | 67177..67401 | + | 225 | WP_060446845 | TraR/DksA C4-type zinc finger protein | - |
| JLH17_RS26185 (STN0717ENT53_P30850) | 67405..67590 | + | 186 | WP_072271147 | hypothetical protein | - |
| JLH17_RS26190 (STN0717ENT53_P30860) | 67574..67978 | + | 405 | WP_060446847 | hypothetical protein | - |
| JLH17_RS26195 (STN0717ENT53_P30870) | 67968..68531 | + | 564 | WP_060446829 | conjugal transfer pilus-stabilizing protein TraP | - |
| JLH17_RS26200 (STN0717ENT53_P30880) | 68524..71154 | + | 2631 | WP_200928253 | type IV secretion system protein TraC | virb4 |
| JLH17_RS26205 (STN0717ENT53_P30890) | 71151..71498 | + | 348 | WP_200928254 | type-F conjugative transfer system protein TrbI | - |
| JLH17_RS26210 (STN0717ENT53_P30900) | 71498..72145 | + | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| JLH17_RS26215 (STN0717ENT53_P30910) | 72148..73134 | + | 987 | WP_032637387 | conjugal transfer pilus assembly protein TraU | traU |
| JLH17_RS26220 (STN0717ENT53_P30920) | 73151..73780 | + | 630 | WP_163310344 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| JLH17_RS26225 (STN0717ENT53_P30930) | 73777..75627 | + | 1851 | WP_200928255 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| JLH17_RS26230 (STN0717ENT53_P30940) | 75665..75853 | + | 189 | WP_125885541 | hypothetical protein | - |
| JLH17_RS26235 (STN0717ENT53_P30950) | 75883..76632 | + | 750 | WP_126336472 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| JLH17_RS26240 (STN0717ENT53_P30960) | 76649..76897 | + | 249 | WP_011901838 | type-F conjugative transfer system pilin chaperone TraQ | - |
| JLH17_RS26245 (STN0717ENT53_P30970) | 76887..77441 | + | 555 | WP_058610060 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| JLH17_RS26250 (STN0717ENT53_P30980) | 77434..77862 | + | 429 | WP_008324122 | hypothetical protein | - |
| JLH17_RS26255 (STN0717ENT53_P30990) | 77849..79219 | + | 1371 | WP_032672666 | conjugal transfer pilus assembly protein TraH | traH |
| JLH17_RS26260 (STN0717ENT53_P31000) | 79219..82080 | + | 2862 | WP_126336474 | conjugal transfer mating-pair stabilization protein TraG | traG |
| JLH17_RS26270 (STN0717ENT53_P31020) | 82660..83385 | + | 726 | WP_060446836 | conjugal transfer complement resistance protein TraT | - |
| JLH17_RS26275 (STN0717ENT53_P31030) | 83756..85984 | + | 2229 | WP_200928256 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 18697 | GenBank | NZ_AP022501 |
| Plasmid name | pSTN0717-53-3 | Incompatibility group | - |
| Plasmid size | 97602 bp | Coordinate of oriT [Strand] | 60805..60861 [-] |
| Host baterium | Enterobacter kobei strain STN0717-53 |
Cargo genes
| Drug resistance gene | aadA5, qacE, sul1, qnrA1, blaSHV-12 |
| Virulence gene | - |
| Metal resistance gene | merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |