Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118260 |
| Name | oriT_pSTW0522-66-2 |
| Organism | Enterobacter roggenkampii strain STW0522-66 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP022467 (3372..3472 [-], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 39..44, 49..54 (CGCACC..GGTGCG) 13..21, 27..35 (AAGTCATTG..CAATGACTT) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
TGTCTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_pSTW0522-66-2
TCCTTGGCGTGGAAGTCATTGTAAATCAATGACTTACGCGCACCGAAAGGTGCGTATTGTCTATAGCCCAGATTTAAGGATACCAACCCGGCTTTTAAGGA
TCCTTGGCGTGGAAGTCATTGTAAATCAATGACTTACGCGCACCGAAAGGTGCGTATTGTCTATAGCCCAGATTTAAGGATACCAACCCGGCTTTTAAGGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file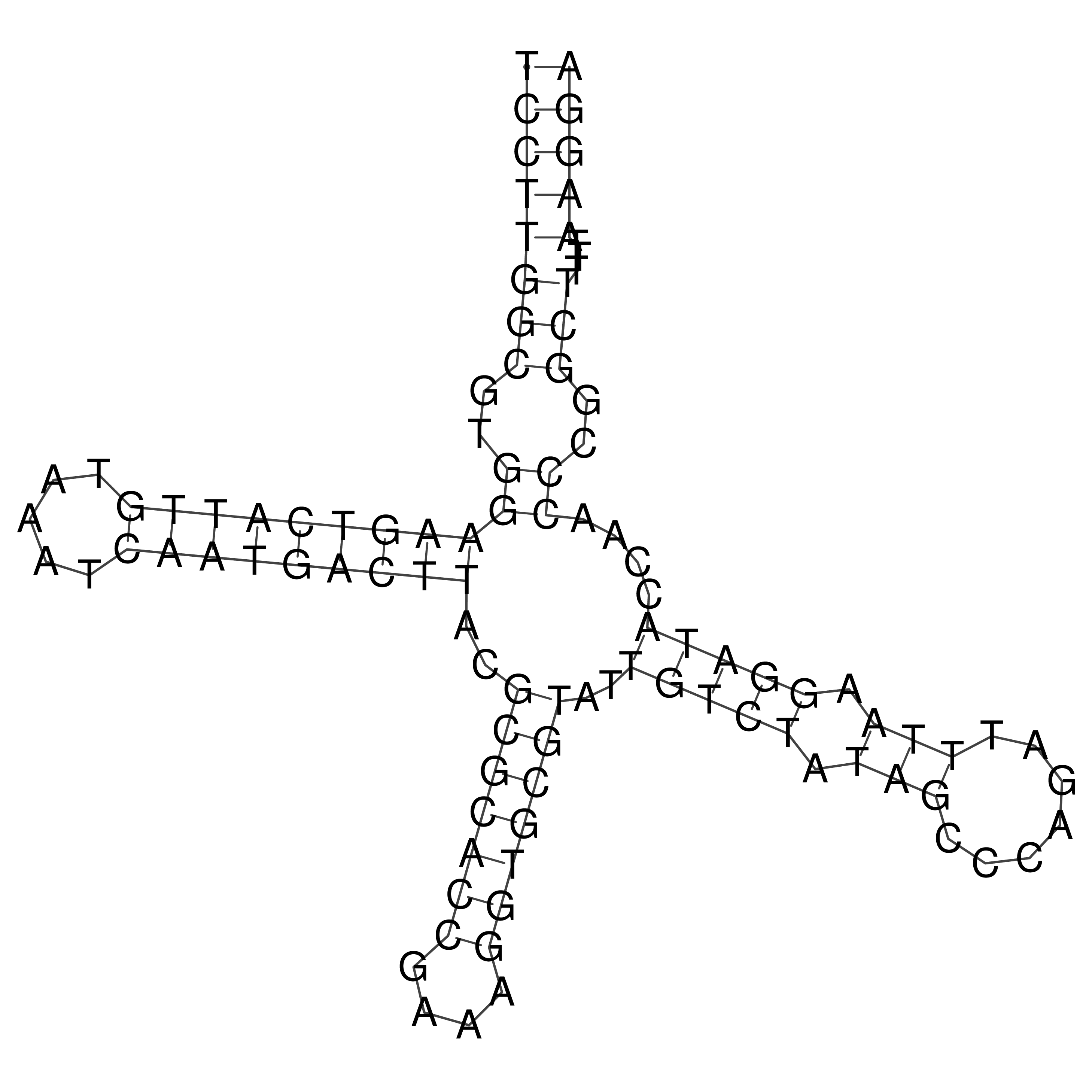
Relaxase
| ID | 11608 | GenBank | WP_200925855 |
| Name | mobF_JLF42_RS25770_pSTW0522-66-2 |
UniProt ID | _ |
| Length | 595 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 595 a.a. Molecular weight: 65781.19 Da Isoelectric Point: 9.6947
>WP_200925855.1 MobF family relaxase [Enterobacter roggenkampii]
MLSHMVLTRQDIGRAASYYEDGADDYYAKDGDASEWQGKGAEELGLSGEVDSKRFRELLAGNIGEGHRIM
RSATRQDSKERIGLDLTFSAPKSVSLQALVAGDAEIIKAHDRAVARTLEQAEARAQARQKIQGKTRIETT
GNLVIGKFRHETSRERDPQLHTHAVILNMTKRSDGQWRALKNDEIVKATRYLGAVYNAELAHELQKLGYQ
LRYGKDGNFDLAHIDRQQIEGFSKRTEQIAEWYAARGLDPNSVSLEQKQAAKVLSRAKKTSVDREALRAE
WQATAKELGIDFSRREWSGREKGGSEKQAHSFMPSDEAAKRAVRYAINHLTERQSVMDERELVDTAMKHA
VGAARLEDIQKELLRQTETGYLIREAPRYRPGGQTGPTDEPGKTRAEWVAELAAKGMKQGAARERVDNAI
KTGGLVPIEPRYTTQTALEREKRILQIERDGRGAVAPVIAAEAARERLASTNLNQGQREAAELIVSAANR
VVGVQGFAGTGKSHMLDTAKQMIEGEGYHVRALAPYGSQVKALRELNVEANTLASFLRAKDKNIDSRTVL
VIDEAGVVPTRLMEQTLKLAEKAGARVVLMGVGSG
MLSHMVLTRQDIGRAASYYEDGADDYYAKDGDASEWQGKGAEELGLSGEVDSKRFRELLAGNIGEGHRIM
RSATRQDSKERIGLDLTFSAPKSVSLQALVAGDAEIIKAHDRAVARTLEQAEARAQARQKIQGKTRIETT
GNLVIGKFRHETSRERDPQLHTHAVILNMTKRSDGQWRALKNDEIVKATRYLGAVYNAELAHELQKLGYQ
LRYGKDGNFDLAHIDRQQIEGFSKRTEQIAEWYAARGLDPNSVSLEQKQAAKVLSRAKKTSVDREALRAE
WQATAKELGIDFSRREWSGREKGGSEKQAHSFMPSDEAAKRAVRYAINHLTERQSVMDERELVDTAMKHA
VGAARLEDIQKELLRQTETGYLIREAPRYRPGGQTGPTDEPGKTRAEWVAELAAKGMKQGAARERVDNAI
KTGGLVPIEPRYTTQTALEREKRILQIERDGRGAVAPVIAAEAARERLASTNLNQGQREAAELIVSAANR
VVGVQGFAGTGKSHMLDTAKQMIEGEGYHVRALAPYGSQVKALRELNVEANTLASFLRAKDKNIDSRTVL
VIDEAGVVPTRLMEQTLKLAEKAGARVVLMGVGSG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6708 | GenBank | WP_012196434 |
| Name | WP_012196434_pSTW0522-66-2 |
UniProt ID | A8R762 |
| Length | 121 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 121 a.a. Molecular weight: 13400.32 Da Isoelectric Point: 4.8464
>WP_012196434.1 MULTISPECIES: hypothetical protein [Pseudomonadota]
MALGDPIQVRLSPEKQALLEDEAARKGKRLATYLRELLESENDLQGELAALRREVVSLHHVIEDLADTGL
RSDQSGPGQNAVQIETLLLLRAIAGPERMKPVKGELKRLGIEVWTPEGKED
MALGDPIQVRLSPEKQALLEDEAARKGKRLATYLRELLESENDLQGELAALRREVVSLHHVIEDLADTGL
RSDQSGPGQNAVQIETLLLLRAIAGPERMKPVKGELKRLGIEVWTPEGKED
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A8R762 |
T4CP
| ID | 13593 | GenBank | WP_012196433 |
| Name | t4cp2_JLF42_RS25765_pSTW0522-66-2 |
UniProt ID | _ |
| Length | 507 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 507 a.a. Molecular weight: 56272.95 Da Isoelectric Point: 9.8647
>WP_012196433.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Pseudomonadota]
MHPDDQRKVSAGIVIVLPLIFWITAVQKTEVLGSPKLLALWELMKLTPQKPILLLSALGGLAVGVLFVWL
LNSVGQGEFGGAPFKRFLRGTRIVSGGKLKRMTREKAKQVTVAGVPMPRDAEPRHLLVNGATGTGKSVLL
RELAYTGLLRGDRMVIVDPNGDMLSKFGRDKDIILNPYDQRTKGWSFFNEIRNDYDWQRYALSVVPRGKT
DEAEEWASYGRLLLRETAKKLALIGTPSMRELFHWTTIATFDDLRGFLEGTLAESLFAGSNEASKALTSA
RFVLSDKLPEHVTMPDGDFSIRSWLEDPNGGNLFITWREDMGPALRPLISAWVDVVCTSILSLPEEPKRR
LWLFIDELASLEKLASLADALTKGRKAGLRVVAGLQSTSQLDDVYGVKEAQTLRASFRSLVVLGGSRTDP
KTNEDMSLSLGEHEVERDRYSKNTGKHHSTGRALERVRERVVMPAEIANLPDLTAYVGFAGNRPIAKVPL
EIKQFANRQPAFVEGTI
MHPDDQRKVSAGIVIVLPLIFWITAVQKTEVLGSPKLLALWELMKLTPQKPILLLSALGGLAVGVLFVWL
LNSVGQGEFGGAPFKRFLRGTRIVSGGKLKRMTREKAKQVTVAGVPMPRDAEPRHLLVNGATGTGKSVLL
RELAYTGLLRGDRMVIVDPNGDMLSKFGRDKDIILNPYDQRTKGWSFFNEIRNDYDWQRYALSVVPRGKT
DEAEEWASYGRLLLRETAKKLALIGTPSMRELFHWTTIATFDDLRGFLEGTLAESLFAGSNEASKALTSA
RFVLSDKLPEHVTMPDGDFSIRSWLEDPNGGNLFITWREDMGPALRPLISAWVDVVCTSILSLPEEPKRR
LWLFIDELASLEKLASLADALTKGRKAGLRVVAGLQSTSQLDDVYGVKEAQTLRASFRSLVVLGGSRTDP
KTNEDMSLSLGEHEVERDRYSKNTGKHHSTGRALERVRERVVMPAEIANLPDLTAYVGFAGNRPIAKVPL
EIKQFANRQPAFVEGTI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3934..17297
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JLF42_RS25730 (STW0522ENT66_P20010) | 102..374 | + | 273 | WP_012443531 | hypothetical protein | - |
| JLF42_RS25735 (STW0522ENT66_P20020) | 390..1043 | + | 654 | WP_012196439 | hypothetical protein | - |
| JLF42_RS25740 (STW0522ENT66_P20030) | 1191..1442 | + | 252 | WP_012196438 | hypothetical protein | - |
| JLF42_RS25745 (STW0522ENT66_P20040) | 1596..1982 | - | 387 | WP_012196437 | hypothetical protein | - |
| JLF42_RS25750 (STW0522ENT66_P20050) | 1996..2691 | - | 696 | WP_012196436 | StbB family protein | - |
| JLF42_RS25755 (STW0522ENT66_P20060) | 2688..3113 | - | 426 | WP_012196435 | hypothetical protein | - |
| JLF42_RS25760 (STW0522ENT66_P20070) | 3567..3932 | + | 366 | WP_012196434 | hypothetical protein | - |
| JLF42_RS25765 (STW0522ENT66_P20080) | 3934..5457 | + | 1524 | WP_012196433 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| JLF42_RS25770 (STW0522ENT66_P20090) | 5457..7244 | + | 1788 | WP_200925855 | MobF family relaxase | - |
| JLF42_RS25775 (STW0522ENT66_P20100) | 7260..10157 | - | 2898 | WP_001553819 | Tn3-like element Tn5403 family transposase | - |
| JLF42_RS25780 (STW0522ENT66_P20110) | 10252..10857 | + | 606 | WP_000509966 | recombinase family protein | - |
| JLF42_RS25785 | 10898..11041 | - | 144 | WP_012196427 | trwH protein | - |
| JLF42_RS25790 (STW0522ENT66_P20120) | 11154..12182 | - | 1029 | WP_012196426 | type IV secretion system protein | virB6 |
| JLF42_RS25795 (STW0522ENT66_P20130) | 12196..12426 | - | 231 | WP_012196425 | EexN family lipoprotein | - |
| JLF42_RS25800 (STW0522ENT66_P20140) | 12423..13112 | - | 690 | WP_012196424 | P-type DNA transfer protein VirB5 | virB5 |
| JLF42_RS25805 (STW0522ENT66_P20150) | 13109..15580 | - | 2472 | WP_012196423 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| JLF42_RS25810 (STW0522ENT66_P20160) | 15583..15897 | - | 315 | WP_012196422 | type IV secretion system protein VirB3 | virB3 |
| JLF42_RS25815 (STW0522ENT66_P20170) | 15910..16215 | - | 306 | WP_031942907 | VirB2 family type IV secretion system major pilin TrwL | virB2 |
| JLF42_RS25820 (STW0522ENT66_P20180) | 16181..16477 | - | 297 | WP_012196420 | KorA family transcriptional regulator | - |
| JLF42_RS25825 (STW0522ENT66_P20190) | 16599..17297 | - | 699 | WP_012443549 | lytic transglycosylase domain-containing protein | virB1 |
| JLF42_RS25830 (STW0522ENT66_P20200) | 17299..17892 | - | 594 | WP_012414173 | exported protein | - |
| JLF42_RS25835 (STW0522ENT66_P20210) | 18029..18316 | + | 288 | WP_012414174 | H-NS family nucleoid-associated regulatory protein | - |
| JLF42_RS25840 (STW0522ENT66_P20220) | 18613..19113 | - | 501 | WP_000376623 | GNAT family N-acetyltransferase | - |
| JLF42_RS25845 (STW0522ENT66_P20230) | 19241..20080 | - | 840 | WP_000259031 | sulfonamide-resistant dihydropteroate synthase Sul1 | - |
| JLF42_RS25850 (STW0522ENT66_P20240) | 20074..20421 | - | 348 | WP_000679427 | quaternary ammonium compound efflux SMR transporter QacE delta 1 | - |
| JLF42_RS25855 (STW0522ENT66_P20250) | 20590..21144 | - | 555 | WP_012695458 | AAC(6')-Ib family aminoglycoside 6'-N-acetyltransferase | - |
| JLF42_RS25860 (STW0522ENT66_P20260) | 21282..22145 | - | 864 | WP_200925854 | carbapenem-hydrolyzing class A beta-lactamase GES-51 | - |
Host bacterium
| ID | 18693 | GenBank | NZ_AP022467 |
| Plasmid name | pSTW0522-66-2 | Incompatibility group | IncW |
| Plasmid size | 33421 bp | Coordinate of oriT [Strand] | 3372..3472 [-] |
| Host baterium | Enterobacter roggenkampii strain STW0522-66 |
Cargo genes
| Drug resistance gene | sul1, qacE, blaGES-5 |
| Virulence gene | trwH1, trwK, trwM |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |