Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118021 |
| Name | oriT_TSDC17.2-1.1|36 |
| Organism | Bacteroides finegoldii strain Bacteroides finegoldii TSDC17.2-1.1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_OY725938 (17436..17506 [-], 71 nt) |
| oriT length | 71 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 71 nt
>oriT_TSDC17.2-1.1|36
CTTATTGGGGAATTTTCAGCGATACGTAGTATTGCGGCTCGGAAAATTCCCTAATAAGCTACGGTATTTTC
CTTATTGGGGAATTTTCAGCGATACGTAGTATTGCGGCTCGGAAAATTCCCTAATAAGCTACGGTATTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file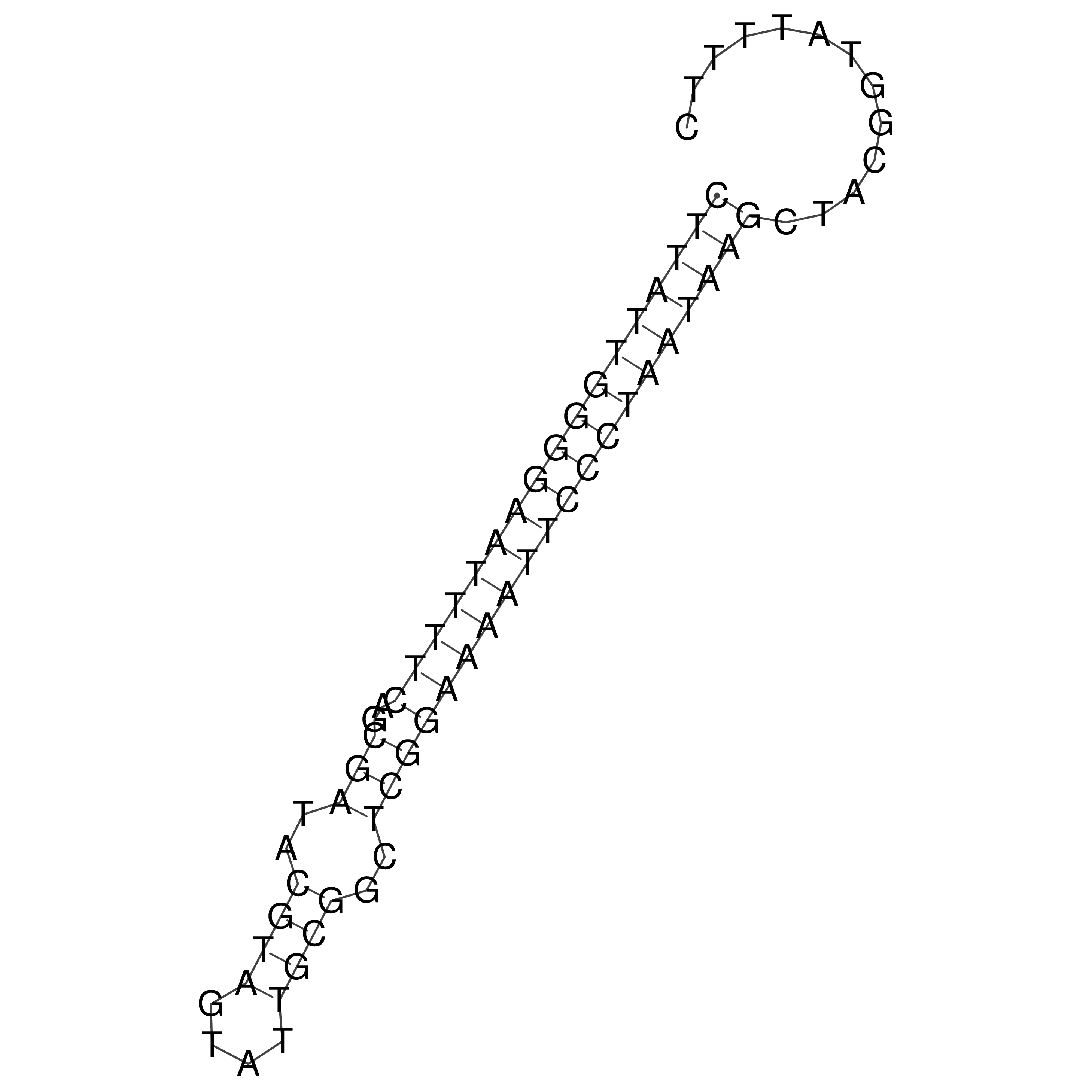
Relaxase
| ID | 11429 | GenBank | WP_008999098 |
| Name | mobV_UHF62_RS14365_TSDC17.2-1.1|36 |
UniProt ID | A0A413X1B7 |
| Length | 455 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 455 a.a. Molecular weight: 52631.91 Da Isoelectric Point: 10.0555
>WP_008999098.1 MULTISPECIES: MobV family relaxase [Bacteroidales]
MGFVVLHMEKAHGSDSGTTAHIERFIIPKNADPTRTHLNRRLIEYPDGIKDRSAAIQQRLEEAGLTRKIG
SNQVRAIRINVSGTHEDMKRIEEEGRLDEWCADNLKYFADTFGKENIVAAHLHRDEETPHIHVTLVPIVK
GERKRRKREEQTKKRYRKKPTDTVRLCADDIMTRLKLKSYQDTYAVAMAKYGLQRGIDGSKARHKSTQQY
YRDIQKLSDDLKAEVVDLQQQKETAQEELRRAKKEIQTEKLKGAATTAAANIAESVGSLFGSNKVKTLER
ENTALHKEVADHEETIEALQDRIQTMQADHSREIREMQQKHGREIADKDTRHKQEISFLKTVIARAAAWF
PYFREMLRIENLCRLVGFDERQTATLVKGKPLEYAGELYSEEHGRKFTTERAGFQVLKDPTDGTKLVLAI
DRKPIAEWFKEQFEKLRQNIRRPIQPQRKGKGFKL
MGFVVLHMEKAHGSDSGTTAHIERFIIPKNADPTRTHLNRRLIEYPDGIKDRSAAIQQRLEEAGLTRKIG
SNQVRAIRINVSGTHEDMKRIEEEGRLDEWCADNLKYFADTFGKENIVAAHLHRDEETPHIHVTLVPIVK
GERKRRKREEQTKKRYRKKPTDTVRLCADDIMTRLKLKSYQDTYAVAMAKYGLQRGIDGSKARHKSTQQY
YRDIQKLSDDLKAEVVDLQQQKETAQEELRRAKKEIQTEKLKGAATTAAANIAESVGSLFGSNKVKTLER
ENTALHKEVADHEETIEALQDRIQTMQADHSREIREMQQKHGREIADKDTRHKQEISFLKTVIARAAAWF
PYFREMLRIENLCRLVGFDERQTATLVKGKPLEYAGELYSEEHGRKFTTERAGFQVLKDPTDGTKLVLAI
DRKPIAEWFKEQFEKLRQNIRRPIQPQRKGKGFKL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A413X1B7 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 115..7012
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| UHF62_RS14250 (BFTSDC17_03178) | 115..558 | + | 444 | WP_255344453 | conjugal transfer protein TraG | virb4 |
| UHF62_RS14255 (BFTSDC17_03179) | 579..1211 | + | 633 | WP_005680097 | DUF4141 domain-containing protein | traI |
| UHF62_RS14260 (BFTSDC17_03180) | 1236..2255 | + | 1020 | WP_008644756 | conjugative transposon protein TraJ | traJ |
| UHF62_RS14265 (BFTSDC17_03181) | 2277..2900 | + | 624 | Protein_3 | conjugative transposon protein TraK | - |
| UHF62_RS14270 (BFTSDC17_03182) | 2913..3173 | + | 261 | WP_005680094 | hypothetical protein | - |
| UHF62_RS14275 (BFTSDC17_03183) | 3173..3361 | + | 189 | WP_005680093 | hypothetical protein | traM |
| UHF62_RS14280 (BFTSDC17_03184) | 3362..4477 | + | 1116 | WP_074706886 | conjugative transposon protein TraM | traM |
| UHF62_RS14285 (BFTSDC17_03185) | 4511..5410 | + | 900 | WP_005680091 | conjugative transposon protein TraN | traN |
| UHF62_RS14290 (BFTSDC17_03186) | 5413..5997 | + | 585 | WP_008644754 | conjugal transfer protein TraO | traO |
| UHF62_RS14295 (BFTSDC17_03187) | 6008..6496 | + | 489 | WP_005680089 | hypothetical protein | - |
| UHF62_RS14300 (BFTSDC17_03188) | 6548..7012 | + | 465 | WP_005680087 | TraQ conjugal transfer family protein | traQ |
| UHF62_RS14305 (BFTSDC17_03189) | 7024..7509 | + | 486 | WP_005680086 | hypothetical protein | - |
| UHF62_RS14310 (BFTSDC17_03190) | 7588..8361 | - | 774 | WP_004308051 | phage antirepressor KilAC domain-containing protein | - |
| UHF62_RS14315 | 8459..8899 | - | 441 | WP_225739787 | hypothetical protein | - |
| UHF62_RS14320 | 8957..9238 | - | 282 | WP_229032698 | DNA cytosine methyltransferase | - |
| UHF62_RS14325 | 9296..9529 | - | 234 | Protein_15 | DNA cytosine methyltransferase | - |
| UHF62_RS14330 (BFTSDC17_03193) | 9629..9976 | - | 348 | WP_005680084 | hypothetical protein | - |
| UHF62_RS14335 (BFTSDC17_03194) | 9989..10309 | - | 321 | WP_005680083 | helix-turn-helix domain-containing protein | - |
| UHF62_RS14340 (BFTSDC17_03195) | 10821..11648 | + | 828 | WP_008644752 | Hachiman antiphage defense system protein HamA | - |
Host bacterium
| ID | 18454 | GenBank | NZ_OY725938 |
| Plasmid name | TSDC17.2-1.1|36 | Incompatibility group | - |
| Plasmid size | 43031 bp | Coordinate of oriT [Strand] | 17436..17506 [-] |
| Host baterium | Bacteroides finegoldii strain Bacteroides finegoldii TSDC17.2-1.1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |