Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 118018 |
| Name | oriT_pJHP9_2 |
| Organism | Enterococcus faecium strain JHP9 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP026569 (53234..53425 [-], 192 nt) |
| oriT length | 192 nt |
| IRs (inverted repeats) | 106..112, 122..128 (ATTTTTT..AAAAAAT) 107..113, 120..126 (TTTTTTG..CAAAAAA) 93..98, 105..110 (AAAATA..TATTTT) 65..71, 75..81 (AGTTGGC..GCCAACT) 41..49, 52..60 (CACCTTCCT..AGGAAGGTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 192 nt
>oriT_pJHP9_2
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCTAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATAGCTTCATATTTTTTGCTATCACAAAAAAATCCATTTTCGACCTATTTTCGGTCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCTAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATAGCTTCATATTTTTTGCTATCACAAAAAAATCCATTTTCGACCTATTTTCGGTCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file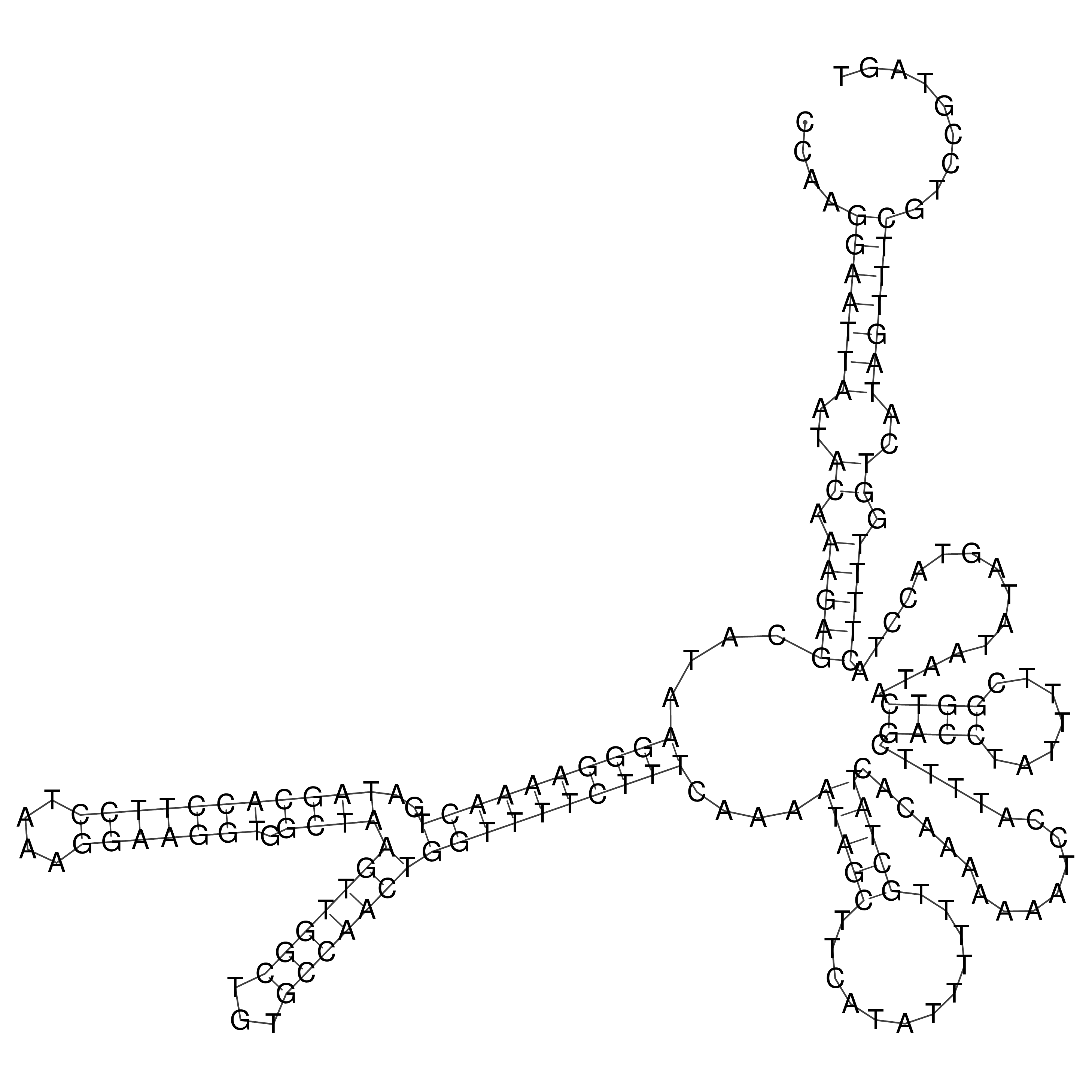
Relaxase
| ID | 11426 | GenBank | WP_265169085 |
| Name | mobL_OQH78_RS16680_pJHP9_2 |
UniProt ID | _ |
| Length | 181 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 181 a.a. Molecular weight: 21355.48 Da Isoelectric Point: 7.3971
>WP_265169085.1 relaxase MobL [Enterococcus faecium]
MEKIEAFSNTKRYITKKDISKIKESVIEAKNNGSVMFQDVISFDNDFLVREGYYNPETNELNENVLYEAT
KSMMGKMQEKEELVDPFWFATIHRNTEHIHIHVTAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFG
SKILDRTNEFEKISKLRKEVPLELKQSVKDSLIQLYVVAKY
MEKIEAFSNTKRYITKKDISKIKESVIEAKNNGSVMFQDVISFDNDFLVREGYYNPETNELNENVLYEAT
KSMMGKMQEKEELVDPFWFATIHRNTEHIHIHVTAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFG
SKILDRTNEFEKISKLRKEVPLELKQSVKDSLIQLYVVAKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13356 | GenBank | WP_265169075 |
| Name | t4cp2_OQH78_RS16610_pJHP9_2 |
UniProt ID | _ |
| Length | 809 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 809 a.a. Molecular weight: 92060.35 Da Isoelectric Point: 8.3857
>WP_265169075.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecium]
MKLVQNNEVKEIKNGFVKNSNKILGKLKKVGSFKKNKYHTGKIEGEDLPKSLQDKLNVPLVFIGVFLGSI
VFLAITYLLNFLKALLVAIGQSESGLLGVEFDPKFSLAYLKPLGDTKQYFMCAVVAVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLEKRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMGTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRKENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGKNYSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYARFIKDLKTRLEYSIINNYQDYLNLSGSQQNN
MKLVQNNEVKEIKNGFVKNSNKILGKLKKVGSFKKNKYHTGKIEGEDLPKSLQDKLNVPLVFIGVFLGSI
VFLAITYLLNFLKALLVAIGQSESGLLGVEFDPKFSLAYLKPLGDTKQYFMCAVVAVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLEKRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMGTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRKENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGKNYSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYARFIKDLKTRLEYSIINNYQDYLNLSGSQQNN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 34405..61756
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OQH78_RS16535 (EfmJHP9_33310) | 29687..31918 | + | 2232 | WP_265169123 | collagen binding domain-containing protein | - |
| OQH78_RS16540 (EfmJHP9_33320) | 32077..32286 | + | 210 | WP_265169068 | hypothetical protein | - |
| OQH78_RS16545 (EfmJHP9_33330) | 32668..33315 | + | 648 | WP_265169070 | hypothetical protein | - |
| OQH78_RS16550 (EfmJHP9_33340) | 33375..33737 | + | 363 | WP_002311845 | hypothetical protein | - |
| OQH78_RS16555 (EfmJHP9_33350) | 33751..33903 | + | 153 | WP_002311846 | hypothetical protein | - |
| OQH78_RS16560 (EfmJHP9_33360) | 33971..34393 | + | 423 | WP_002311847 | hypothetical protein | - |
| OQH78_RS16565 (EfmJHP9_33370) | 34405..35820 | + | 1416 | WP_002311848 | CpaF/VirB11 family protein | virB11 |
| OQH78_RS16570 (EfmJHP9_33380) | 35837..36652 | + | 816 | WP_002313315 | hypothetical protein | - |
| OQH78_RS16575 (EfmJHP9_33390) | 36652..37449 | + | 798 | WP_002311850 | hypothetical protein | - |
| OQH78_RS16580 (EfmJHP9_33400) | 37467..37829 | + | 363 | WP_002311851 | DUF4320 family protein | - |
| OQH78_RS16585 (EfmJHP9_33410) | 37843..37989 | + | 147 | WP_002326866 | hypothetical protein | - |
| OQH78_RS16590 (EfmJHP9_33420) | 38001..38222 | + | 222 | WP_002326867 | hypothetical protein | - |
| OQH78_RS16595 (EfmJHP9_33430) | 38241..38516 | + | 276 | WP_002311852 | TrbC/VirB2 family protein | virB2 |
| OQH78_RS16600 (EfmJHP9_33440) | 38625..40451 | + | 1827 | WP_002311853 | hypothetical protein | - |
| OQH78_RS16605 (EfmJHP9_33450) | 40464..41105 | + | 642 | WP_002354483 | hypothetical protein | - |
| OQH78_RS16610 (EfmJHP9_33460) | 41193..43622 | + | 2430 | WP_265169075 | VirD4-like conjugal transfer protein, CD1115 family | - |
| OQH78_RS16615 (EfmJHP9_33470) | 43692..44051 | + | 360 | WP_265169076 | hypothetical protein | - |
| OQH78_RS16620 (EfmJHP9_33490) | 44102..46739 | + | 2638 | Protein_50 | hypothetical protein | - |
| OQH78_RS16625 (EfmJHP9_33500) | 46787..47098 | + | 312 | WP_002317413 | DUF5592 family protein | - |
| OQH78_RS16630 (EfmJHP9_33510) | 47095..47712 | + | 618 | WP_265169077 | hypothetical protein | virb4 |
| OQH78_RS16635 (EfmJHP9_33520) | 47728..48195 | + | 468 | WP_265169078 | hypothetical protein | - |
| OQH78_RS16640 (EfmJHP9_33530) | 48211..50166 | + | 1956 | WP_002311860 | VirB4 family type IV secretion system protein | virb4 |
| OQH78_RS16645 (EfmJHP9_33540) | 50188..51324 | + | 1137 | WP_002311861 | bifunctional lytic transglycosylase/C40 family peptidase | orf14 |
| OQH78_RS16650 (EfmJHP9_33550) | 51339..51989 | + | 651 | WP_002311862 | cell division protein FtsL | - |
| OQH78_RS16655 (EfmJHP9_33560) | 52003..52296 | + | 294 | WP_265169079 | hypothetical protein | - |
| OQH78_RS16660 (EfmJHP9_33570) | 52933..53163 | + | 231 | WP_002311864 | hypothetical protein | - |
| OQH78_RS16665 (EfmJHP9_33580) | 53514..53810 | + | 297 | WP_002311865 | hypothetical protein | - |
| OQH78_RS16670 (EfmJHP9_33590) | 53807..54274 | + | 468 | WP_002311867 | hypothetical protein | - |
| OQH78_RS16675 (EfmJHP9_33600) | 54300..54506 | + | 207 | WP_265169083 | hypothetical protein | - |
| OQH78_RS16680 (EfmJHP9_33610) | 54533..55078 | + | 546 | WP_265169085 | relaxase MobL | - |
| OQH78_RS16685 (EfmJHP9_33620) | 55111..55422 | + | 312 | WP_265169087 | hypothetical protein | - |
| OQH78_RS16690 (EfmJHP9_33630) | 55449..55652 | + | 204 | WP_265169089 | hypothetical protein | - |
| OQH78_RS16695 (EfmJHP9_33640) | 55750..56079 | + | 330 | WP_002311869 | hypothetical protein | - |
| OQH78_RS16700 (EfmJHP9_33650) | 56081..56344 | + | 264 | WP_002311870 | hypothetical protein | - |
| OQH78_RS16705 (EfmJHP9_33660) | 56519..57457 | + | 939 | WP_265169092 | hypothetical protein | - |
| OQH78_RS16710 (EfmJHP9_33670) | 57539..59365 | + | 1827 | WP_002311872 | ArdC-like ssDNA-binding domain-containing protein | - |
| OQH78_RS16715 (EfmJHP9_33680) | 59373..60446 | + | 1074 | WP_002311873 | hypothetical protein | - |
| OQH78_RS16720 (EfmJHP9_33690) | 60455..60715 | + | 261 | WP_002311874 | hypothetical protein | - |
| OQH78_RS16725 (EfmJHP9_33700) | 60752..61756 | + | 1005 | WP_002311875 | DUF3991 domain-containing protein | traP |
| OQH78_RS16730 (EfmJHP9_33710) | 61781..62137 | + | 357 | WP_002311876 | hypothetical protein | - |
| OQH78_RS16735 (EfmJHP9_33720) | 62299..62490 | + | 192 | WP_002311877 | hypothetical protein | - |
| OQH78_RS16740 (EfmJHP9_33730) | 62599..62817 | + | 219 | WP_002311878 | hypothetical protein | - |
| OQH78_RS16745 (EfmJHP9_33740) | 62854..63171 | + | 318 | WP_002311879 | hypothetical protein | - |
| OQH78_RS16750 (EfmJHP9_33750) | 63182..63379 | + | 198 | WP_002311880 | hypothetical protein | - |
Host bacterium
| ID | 18451 | GenBank | NZ_AP026569 |
| Plasmid name | pJHP9_2 | Incompatibility group | - |
| Plasmid size | 63729 bp | Coordinate of oriT [Strand] | 53234..53425 [-] |
| Host baterium | Enterococcus faecium strain JHP9 |
Cargo genes
| Drug resistance gene | aac(6')-aph(2''), dfrG |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |