Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117993 |
| Name | oriT_p5D_Hdefensa-2 |
| Organism | Candidatus Hamiltonella defensa strain 5D |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021665 (8103..8181 [-], 79 nt) |
| oriT length | 79 nt |
| IRs (inverted repeats) | 23..30, 33..40 (CCGCTTGC..GCAAGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 79 nt
>oriT_p5D_Hdefensa-2
GGACAAGATATGTTTTGAAGCACCGCTTGCAAGCAAGCGGCCCTTCAAAACTCTTGGTCAGGGCTTCGCCCCGACACCC
GGACAAGATATGTTTTGAAGCACCGCTTGCAAGCAAGCGGCCCTTCAAAACTCTTGGTCAGGGCTTCGCCCCGACACCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file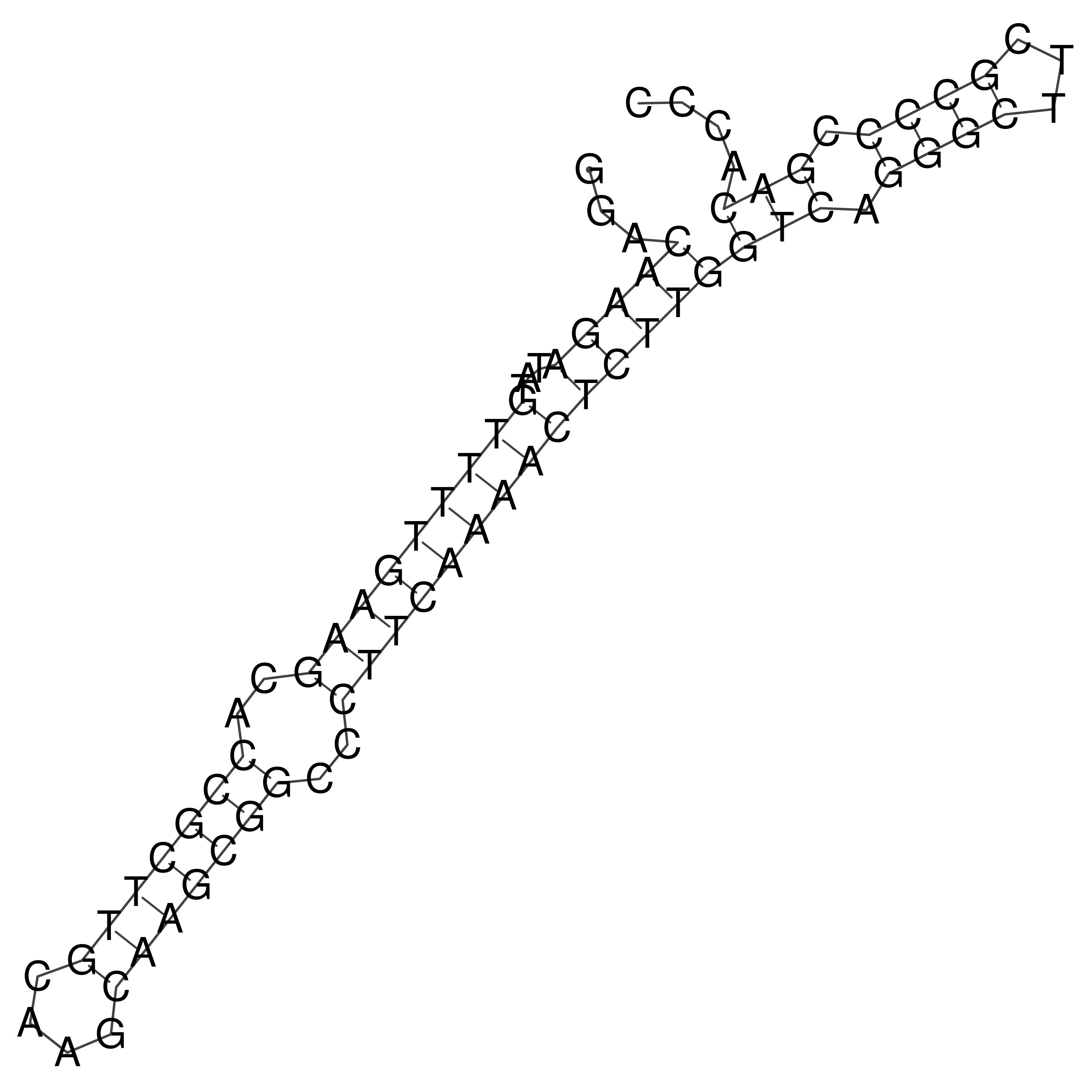
Relaxase
| ID | 11414 | GenBank | WP_095034927 |
| Name | traI_CCS40_RS10760_p5D_Hdefensa-2 |
UniProt ID | _ |
| Length | 627 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 627 a.a. Molecular weight: 72459.28 Da Isoelectric Point: 9.0352
>WP_095034927.1 TraI/MobA(P) family conjugative relaxase [Candidatus Hamiltonella defensa]
MIVGKAKKRRDGKSSFLTLLAYTTTRDDHHHDDVIDPETQKARLSQSKEAIFERLMGYIHRHGDADKSLI
TERFPDGRHQVLFDQVLCETNTLSVATAAVEMQAVALKNTRCKDPVYHFFLSWPETDSPTVEQIFESARH
SLKALGMSDHQYVTAIHRDTDHLHCHVAANRIHPVTYKVADDAYDISKLHKASREMELKYGWTRTNGCHV
INENNRIVRSCSKEKSMPDDAKKLEYYSDQESLYGYAVRECRSEISEILKADSIYWERIHAVLIRAGLEL
KKKGAGLAIYHRAHPEQTPLKASRLHPDLTLSHLEPRAGPFESSPKVDTYDKDRLVFGYVIEKSYDRRLH
ARDHSARHERRLARAEAREELKLRYQHDKKEAKCPSFDAKNRFRTLSMTFRFRRAHVCVAVRDPLMRKLA
WHVLAFEREKAMAQLRLQLKQERAHWHRAPENRRLSYRLWVEQQALKGDKAAISQLRGWAYQAKRDERTA
YLSDTVIECAVSDDIAPVELKGYTHHIHRDGAILYKKEGVTQMIDRGETIEMARPFENEGDNMVAGLRLA
EQKSGEKRVFSGPREYVEKACSLVRDLNEMGETSLTLTDSLQQKRVCEMRPQDKPAPISFDSPNLTP
MIVGKAKKRRDGKSSFLTLLAYTTTRDDHHHDDVIDPETQKARLSQSKEAIFERLMGYIHRHGDADKSLI
TERFPDGRHQVLFDQVLCETNTLSVATAAVEMQAVALKNTRCKDPVYHFFLSWPETDSPTVEQIFESARH
SLKALGMSDHQYVTAIHRDTDHLHCHVAANRIHPVTYKVADDAYDISKLHKASREMELKYGWTRTNGCHV
INENNRIVRSCSKEKSMPDDAKKLEYYSDQESLYGYAVRECRSEISEILKADSIYWERIHAVLIRAGLEL
KKKGAGLAIYHRAHPEQTPLKASRLHPDLTLSHLEPRAGPFESSPKVDTYDKDRLVFGYVIEKSYDRRLH
ARDHSARHERRLARAEAREELKLRYQHDKKEAKCPSFDAKNRFRTLSMTFRFRRAHVCVAVRDPLMRKLA
WHVLAFEREKAMAQLRLQLKQERAHWHRAPENRRLSYRLWVEQQALKGDKAAISQLRGWAYQAKRDERTA
YLSDTVIECAVSDDIAPVELKGYTHHIHRDGAILYKKEGVTQMIDRGETIEMARPFENEGDNMVAGLRLA
EQKSGEKRVFSGPREYVEKACSLVRDLNEMGETSLTLTDSLQQKRVCEMRPQDKPAPISFDSPNLTP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13339 | GenBank | WP_187356329 |
| Name | trbC_CCS40_RS11040_p5D_Hdefensa-2 |
UniProt ID | _ |
| Length | 645 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 645 a.a. Molecular weight: 73110.36 Da Isoelectric Point: 7.4673
>WP_187356329.1 F-type conjugative transfer protein TrbC [Candidatus Hamiltonella defensa]
MIALVLGCLWPVTLLIGLPACVILLIIFCDRPFRMPIRLPTDIGGPDLTTEREGFKARKGWSGLFSFITR
TRHCEQAAGILCLGYARGKSLARELWLNLDDALRHIQLIATTGAGKTEAILSVYLNALCWGRGMCLSDGK
AENRLAFAVWSLARRFGREDDVYILNFLTAGNSKFSDLMKDNKSRPQSNSINLFANASETFIIQLMDSLL
PKVGSNETGWQEKAKAMIAALIYALCYKREKDGLFLSQRVIQDYLPLRKIVTLYQEAKKNHWHEEGYKPL
EHYLNTLAGFDMALIDSPLEWSKTVWEQHGFLIQQFTRMLSLFNDIYGHVFPQGAGDIDLKDVLHNDRIL
VVLIPALELSNPEAVTLGKLYISGLRMTLSQDLGGQLEGKREDVLLSQKFAQKFPYPIIFDELGAYFASG
LDKMAAQMRSLQYMLMIAAQDVQSMMDKSMREFFTVSANQRTKWFMALEDAQDTFNLIRSAAGKGYYSEL
SSVKRKGSLSGSRYEDADTHHIRERDNIELTELKALNKGEGVIVFQDAVVRSAAIFIPDEEKLSSKLPLR
INRFIELKRPDFDALCEVVPELRHKGAAFKQEVQGILQRLKTADTQENKGTLNVRLFDKTLHHLCEITQN
LDHRNDLSYTPRSEE
MIALVLGCLWPVTLLIGLPACVILLIIFCDRPFRMPIRLPTDIGGPDLTTEREGFKARKGWSGLFSFITR
TRHCEQAAGILCLGYARGKSLARELWLNLDDALRHIQLIATTGAGKTEAILSVYLNALCWGRGMCLSDGK
AENRLAFAVWSLARRFGREDDVYILNFLTAGNSKFSDLMKDNKSRPQSNSINLFANASETFIIQLMDSLL
PKVGSNETGWQEKAKAMIAALIYALCYKREKDGLFLSQRVIQDYLPLRKIVTLYQEAKKNHWHEEGYKPL
EHYLNTLAGFDMALIDSPLEWSKTVWEQHGFLIQQFTRMLSLFNDIYGHVFPQGAGDIDLKDVLHNDRIL
VVLIPALELSNPEAVTLGKLYISGLRMTLSQDLGGQLEGKREDVLLSQKFAQKFPYPIIFDELGAYFASG
LDKMAAQMRSLQYMLMIAAQDVQSMMDKSMREFFTVSANQRTKWFMALEDAQDTFNLIRSAAGKGYYSEL
SSVKRKGSLSGSRYEDADTHHIRERDNIELTELKALNKGEGVIVFQDAVVRSAAIFIPDEEKLSSKLPLR
INRFIELKRPDFDALCEVVPELRHKGAAFKQEVQGILQRLKTADTQENKGTLNVRLFDKTLHHLCEITQN
LDHRNDLSYTPRSEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 27295..54943
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CCS40_RS10860 (CCS40_10900) | 22517..24217 | + | 1701 | WP_095034941 | PilN family type IVB pilus formation outer membrane protein | - |
| CCS40_RS10865 (CCS40_10905) | 24229..24567 | - | 339 | WP_095034942 | helix-turn-helix domain-containing protein | - |
| CCS40_RS10870 (CCS40_10910) | 24548..24928 | - | 381 | WP_148121498 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| CCS40_RS10875 (CCS40_10915) | 24992..26302 | + | 1311 | WP_148121499 | type 4b pilus protein PilO2 | - |
| CCS40_RS10880 (CCS40_10920) | 26289..26699 | + | 411 | WP_095034943 | type IV pilus biogenesis protein PilP | - |
| CCS40_RS12690 | 26696..27298 | + | 603 | WP_235655681 | hypothetical protein | - |
| CCS40_RS12695 | 27295..27888 | + | 594 | WP_235655682 | ATPase, T2SS/T4P/T4SS family | virB11 |
| CCS40_RS12700 | 27963..28241 | + | 279 | WP_235655683 | hypothetical protein | - |
| CCS40_RS12705 | 28228..28782 | + | 555 | WP_235655684 | type II secretion system F family protein | - |
| CCS40_RS12710 | 28776..29291 | + | 516 | WP_235655676 | type II secretion system F family protein | - |
| CCS40_RS10895 (CCS40_10935) | 29303..29812 | + | 510 | WP_095034945 | type 4 pilus major pilin | - |
| CCS40_RS10900 (CCS40_10940) | 30065..31300 | + | 1236 | WP_095034946 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| CCS40_RS10905 (CCS40_10945) | 31339..31770 | + | 432 | WP_095034947 | type IV pilus biogenesis protein PilM | - |
| CCS40_RS12715 | 31776..32063 | + | 288 | WP_235655677 | hypothetical protein | - |
| CCS40_RS12720 | 32006..32395 | + | 390 | WP_234813502 | prepilin peptidase | - |
| CCS40_RS10915 (CCS40_10955) | 32438..32908 | + | 471 | WP_095034948 | lytic transglycosylase domain-containing protein | virB1 |
| CCS40_RS10920 (CCS40_10960) | 32962..33405 | + | 444 | WP_095034949 | DotD/TraH family lipoprotein | - |
| CCS40_RS10925 (CCS40_10965) | 33402..34210 | + | 809 | Protein_47 | type IV secretion system DotC family protein | - |
| CCS40_RS10930 (CCS40_10970) | 34197..35363 | + | 1167 | WP_148121500 | plasmid transfer ATPase TraJ | virB11 |
| CCS40_RS10935 (CCS40_10975) | 35425..35685 | + | 261 | WP_015873087 | IcmT/TraK family protein | traK |
| CCS40_RS10940 (CCS40_10980) | 35700..37013 | + | 1314 | WP_148121501 | DUF5710 domain-containing protein | - |
| CCS40_RS10945 (CCS40_10985) | 37022..37795 | + | 774 | WP_148121502 | LPD7 domain-containing protein | - |
| CCS40_RS10950 (CCS40_10990) | 37795..38340 | + | 546 | WP_148121503 | hypothetical protein | - |
| CCS40_RS10955 (CCS40_10995) | 38437..38868 | + | 432 | WP_095034952 | conjugal transfer protein TraL | traL |
| CCS40_RS12725 | 38893..39120 | + | 228 | WP_234813663 | hypothetical protein | traM |
| CCS40_RS10960 (CCS40_11000) | 39117..39563 | + | 447 | WP_234813664 | DotI/IcmL/TraM family protein | traM |
| CCS40_RS10965 (CCS40_11005) | 39602..40567 | + | 966 | WP_234811143 | DotH/IcmK family type IV secretion protein | traN |
| CCS40_RS10970 (CCS40_11010) | 40571..41590 | + | 1020 | WP_234813665 | conjugal transfer protein TraO | traO |
| CCS40_RS12975 | 41611..41742 | + | 132 | WP_268877281 | hypothetical protein | traO |
| CCS40_RS10975 (CCS40_11015) | 41885..42490 | + | 606 | WP_095034953 | conjugal transfer protein TraP | traP |
| CCS40_RS10980 (CCS40_11020) | 42504..43043 | + | 540 | WP_095034954 | conjugal transfer protein TraQ | traQ |
| CCS40_RS10985 (CCS40_11025) | 43101..43496 | + | 396 | WP_095034955 | DUF6750 family protein | traR |
| CCS40_RS10990 (CCS40_11030) | 43526..44128 | + | 603 | WP_148121504 | TraT conjugal transfer protein | traT |
| CCS40_RS10995 (CCS40_11035) | 44227..47297 | + | 3071 | Protein_63 | ATP-binding protein | - |
| CCS40_RS11000 (CCS40_11040) | 47342..48523 | + | 1182 | WP_234813666 | conjugal transfer protein TraW | traW |
| CCS40_RS11005 (CCS40_11045) | 48520..49068 | + | 549 | WP_148121505 | conjugal transfer protein TraX | - |
| CCS40_RS11010 (CCS40_11050) | 49153..51300 | + | 2148 | WP_095034959 | DotA/TraY family protein | traY |
| CCS40_RS11015 (CCS40_11055) | 51375..52013 | + | 639 | WP_095034960 | plasmid IncI1-type surface exclusion protein ExcA | - |
| CCS40_RS11020 (CCS40_11060) | 52058..52363 | - | 306 | WP_095034961 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| CCS40_RS11025 (CCS40_11065) | 52350..52643 | - | 294 | WP_095034962 | CopG family ribbon-helix-helix protein | - |
| CCS40_RS11030 (CCS40_11070) | 52822..53931 | + | 1110 | WP_095034963 | conjugal transfer protein TrbA | trbA |
| CCS40_RS11035 (CCS40_11075) | 54008..54943 | + | 936 | WP_148121506 | DsbC family protein | trbB |
| CCS40_RS11040 | 55043..56980 | + | 1938 | WP_187356329 | F-type conjugative transfer protein TrbC | - |
| CCS40_RS11050 (CCS40_11090) | 57168..57368 | - | 201 | WP_095034965 | hypothetical protein | - |
Host bacterium
| ID | 18426 | GenBank | NZ_CP021665 |
| Plasmid name | p5D_Hdefensa-2 | Incompatibility group | - |
| Plasmid size | 57467 bp | Coordinate of oriT [Strand] | 8103..8181 [-] |
| Host baterium | Candidatus Hamiltonella defensa strain 5D |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |