Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117978 |
| Name | oriT_FDAARGOS_1334|unnamed2 |
| Organism | Klebsiella oxytoca strain FDAARGOS_1334 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP069928 (4428..4551 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 92..99, 113..120 (ATAATGTA..TACATTAT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTATTTGAATCATTAACTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATAAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file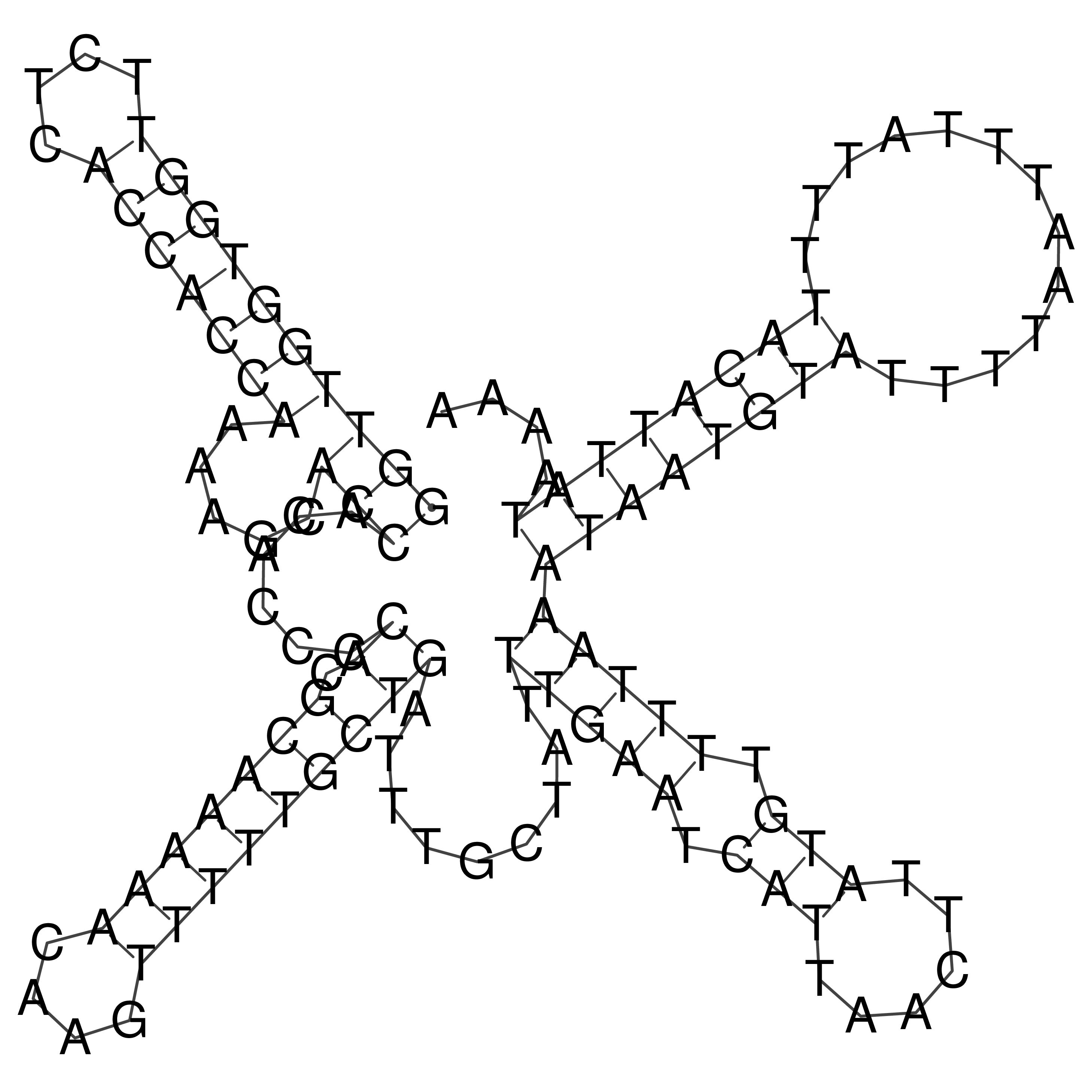
Relaxase
| ID | 11404 | GenBank | WP_204724570 |
| Name | traI_I6K64_RS29125_FDAARGOS_1334|unnamed2 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191793.67 Da Isoelectric Point: 5.9591
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGKGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQDPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEGASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRTEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMVFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYDRLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTEMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWEKNPDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAQRLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRSAISERETALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6555 | GenBank | WP_001151564 |
| Name | WP_001151564_FDAARGOS_1334|unnamed2 |
UniProt ID | A0AA36PD43 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14446.39 Da Isoelectric Point: 4.7197
MAKVQAYVSDEIVYKINKIVERRRAEGAKSTDVSFSSISTMLLELGLRVYEAQMERKESAFNQAEFNKVL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIRDKVSSEMERFFPENDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6556 | GenBank | WP_001254388 |
| Name | WP_001254388_FDAARGOS_1334|unnamed2 |
UniProt ID | W9A0Z2 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9072.20 Da Isoelectric Point: 10.2071
MRRRNARGGISRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVIEENES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W9A0Z2 |
T4CP
| ID | 13326 | GenBank | WP_001064239 |
| Name | traC_I6K64_RS29030_FDAARGOS_1334|unnamed2 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99485.47 Da Isoelectric Point: 6.6265
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASFHGAKITTQTVDAQAFIEIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKKQARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRTLKKLNVIDEGWR
LLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLFPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRREGM
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13327 | GenBank | WP_000009360 |
| Name | traD_I6K64_RS29120_FDAARGOS_1334|unnamed2 |
UniProt ID | _ |
| Length | 732 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 732 a.a. Molecular weight: 83294.06 Da Isoelectric Point: 5.1048
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVSTGKSEVIRRLANYAR
KRGDMVVIYDRSCEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNVANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVVSMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVLNTRAFFRSPSHQIA
EFAAGEIGEKEHLKASLQYSYGADPVRDGISTGKEMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPVSPAINDKKSDSGVNIPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQEN
HPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3856..26520
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| I6K64_RS28935 (I6K64_28930) | 1..257 | + | 257 | WP_149020673 | conjugation system SOS inhibitor PsiB | - |
| I6K64_RS28940 (I6K64_28935) | 254..1016 | + | 763 | Protein_1 | plasmid SOS inhibition protein A | - |
| I6K64_RS29685 | 1195..1344 | + | 150 | Protein_2 | plasmid maintenance protein Mok | - |
| I6K64_RS28945 (I6K64_28940) | 1286..1411 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| I6K64_RS29690 | 1631..1861 | + | 231 | WP_071587244 | hypothetical protein | - |
| I6K64_RS29695 | 1859..2032 | - | 174 | Protein_5 | hypothetical protein | - |
| I6K64_RS28950 (I6K64_28945) | 2330..2617 | + | 288 | WP_000107537 | hypothetical protein | - |
| I6K64_RS28955 (I6K64_28950) | 2738..3559 | + | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| I6K64_RS28960 (I6K64_28955) | 3856..4503 | - | 648 | WP_000614282 | transglycosylase SLT domain-containing protein | virB1 |
| I6K64_RS28965 (I6K64_28960) | 4789..5172 | + | 384 | WP_001151564 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| I6K64_RS28970 (I6K64_28965) | 5366..6052 | + | 687 | WP_000332487 | PAS domain-containing protein | - |
| I6K64_RS28975 (I6K64_28970) | 6146..6373 | + | 228 | WP_001254388 | conjugal transfer relaxosome protein TraY | - |
| I6K64_RS28980 (I6K64_28975) | 6407..6769 | + | 363 | WP_001098998 | type IV conjugative transfer system pilin TraA | - |
| I6K64_RS28985 (I6K64_28980) | 6774..7085 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| I6K64_RS28990 (I6K64_28985) | 7107..7673 | + | 567 | WP_000399804 | type IV conjugative transfer system protein TraE | traE |
| I6K64_RS28995 (I6K64_28990) | 7660..8388 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| I6K64_RS29000 (I6K64_28995) | 8388..9815 | + | 1428 | WP_000146638 | F-type conjugal transfer pilus assembly protein TraB | traB |
| I6K64_RS29005 (I6K64_29000) | 9805..10389 | + | 585 | WP_000002795 | conjugal transfer pilus-stabilizing protein TraP | - |
| I6K64_RS29010 (I6K64_29005) | 10376..10696 | + | 321 | WP_001057302 | conjugal transfer protein TrbD | virb4 |
| I6K64_RS29015 (I6K64_29010) | 10689..10940 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| I6K64_RS29020 (I6K64_29015) | 10937..11452 | + | 516 | WP_000809893 | type IV conjugative transfer system lipoprotein TraV | traV |
| I6K64_RS29025 (I6K64_29020) | 11587..11808 | + | 222 | WP_001278978 | conjugal transfer protein TraR | - |
| I6K64_RS29030 (I6K64_29025) | 11968..14595 | + | 2628 | WP_001064239 | type IV secretion system protein TraC | virb4 |
| I6K64_RS29035 (I6K64_29030) | 14592..14978 | + | 387 | WP_000214082 | type-F conjugative transfer system protein TrbI | - |
| I6K64_RS29040 (I6K64_29035) | 14975..15607 | + | 633 | WP_001203717 | type-F conjugative transfer system protein TraW | traW |
| I6K64_RS29045 (I6K64_29040) | 15604..16596 | + | 993 | WP_204724569 | conjugal transfer pilus assembly protein TraU | traU |
| I6K64_RS29050 (I6K64_29045) | 16626..16931 | + | 306 | WP_000224411 | hypothetical protein | - |
| I6K64_RS29055 (I6K64_29050) | 16940..17578 | + | 639 | WP_001080257 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| I6K64_RS29060 (I6K64_29055) | 17575..19383 | + | 1809 | WP_000821863 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| I6K64_RS29065 (I6K64_29060) | 19410..19667 | + | 258 | WP_000864320 | conjugal transfer protein TrbE | - |
| I6K64_RS29070 (I6K64_29065) | 19654..20403 | + | 750 | WP_001336806 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| I6K64_RS29075 (I6K64_29070) | 20419..20757 | + | 339 | WP_001287905 | conjugal transfer protein TrbA | - |
| I6K64_RS29080 (I6K64_29075) | 20884..21168 | + | 285 | WP_001448202 | type-F conjugative transfer system pilin chaperone TraQ | - |
| I6K64_RS29085 (I6K64_29080) | 21155..21700 | + | 546 | WP_000059824 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| I6K64_RS29090 (I6K64_29085) | 21630..21971 | + | 342 | WP_001448115 | P-type conjugative transfer protein TrbJ | - |
| I6K64_RS29095 (I6K64_29090) | 21952..22344 | + | 393 | WP_000660703 | F-type conjugal transfer protein TrbF | - |
| I6K64_RS29100 (I6K64_29095) | 22331..23704 | + | 1374 | WP_001137364 | conjugal transfer pilus assembly protein TraH | traH |
| I6K64_RS29105 (I6K64_29100) | 23701..26520 | + | 2820 | WP_001007057 | conjugal transfer mating-pair stabilization protein TraG | traG |
| I6K64_RS29110 (I6K64_29105) | 26539..27036 | + | 498 | WP_000605861 | hypothetical protein | - |
| I6K64_RS29115 (I6K64_29110) | 27068..27799 | + | 732 | WP_000850422 | conjugal transfer complement resistance protein TraT | - |
| I6K64_RS29120 (I6K64_29115) | 28052..30250 | + | 2199 | WP_000009360 | type IV conjugative transfer system coupling protein TraD | - |
Host bacterium
| ID | 18411 | GenBank | NZ_CP069928 |
| Plasmid name | FDAARGOS_1334|unnamed2 | Incompatibility group | IncFII |
| Plasmid size | 45379 bp | Coordinate of oriT [Strand] | 4428..4551 [+] |
| Host baterium | Klebsiella oxytoca strain FDAARGOS_1334 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |