Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117900 |
| Name | oriT_pTET32A |
| Organism | Raoultella ornithinolytica strain TET32 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP067414 (65378..65472 [+], 95 nt) |
| oriT length | 95 nt |
| IRs (inverted repeats) | 73..78, 85..90 (AAAAAA..TTTTTT) 73..78, 84..89 (AAAAAA..TTTTTT) 27..34, 37..44 (AGCGTGAT..ATCACGCT) 13..19, 31..37 (TAAATCA..TGATTTA) |
| Location of nic site | 55..56 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 95 nt
>oriT_pTET32A
TTTTTTTTCTTTTAAATCAGTGAGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTTGGTAG
TTTTTTTTCTTTTAAATCAGTGAGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file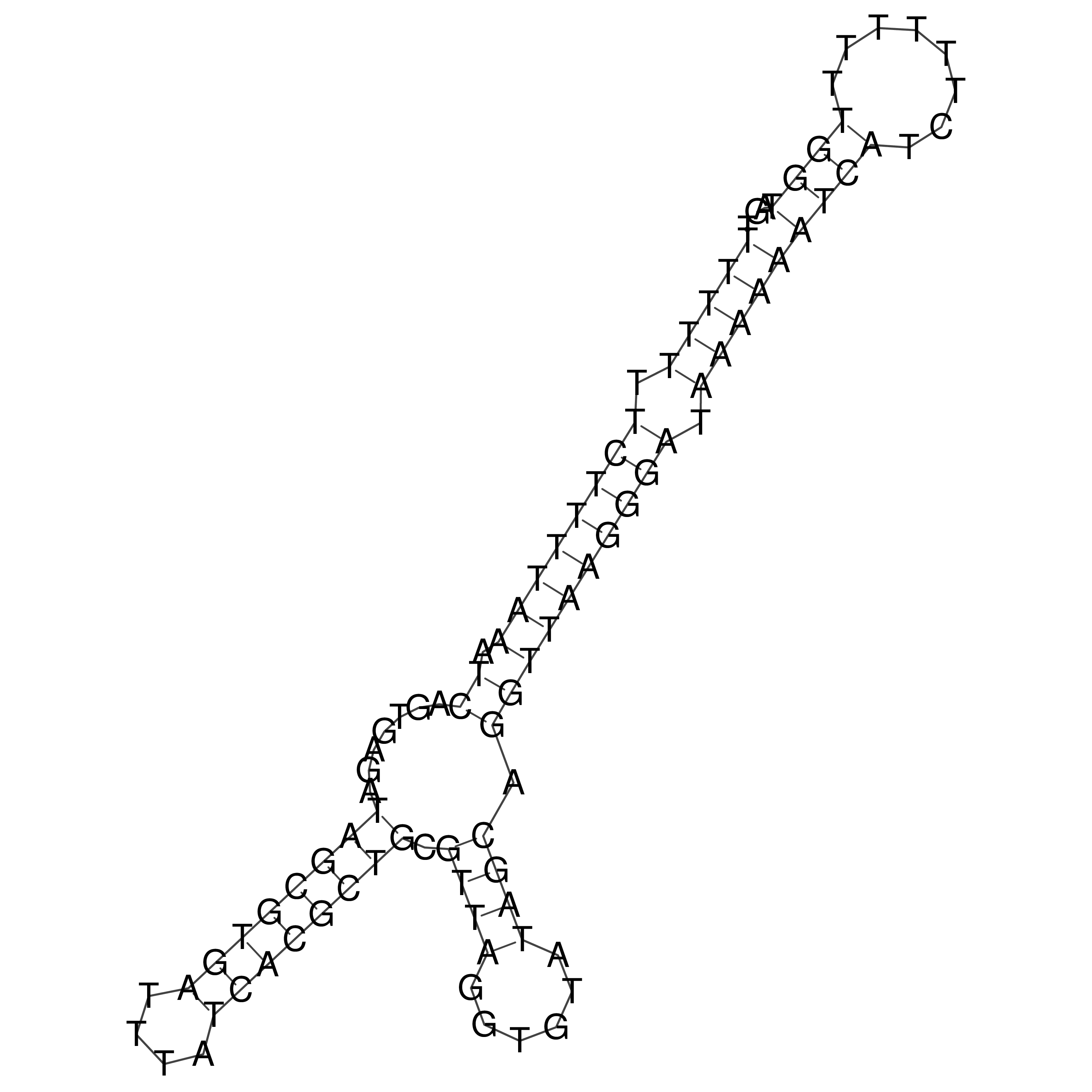
Host bacterium
| ID | 18333 | GenBank | NZ_CP067414 |
| Plasmid name | pTET32A | Incompatibility group | IncR |
| Plasmid size | 68533 bp | Coordinate of oriT [Strand] | 65378..65472 [+] |
| Host baterium | Raoultella ornithinolytica strain TET32 |
Cargo genes
| Drug resistance gene | floR, catA2, tet(D), aac(6')-Ib-cr, ARR-3, dfrA27, aadA16, qacE, sul1, qnrB6 |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merC, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |