Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117823 |
| Name | oriT_unitig_2_pilon |
| Organism | Citrobacter koseri strain AR_0025 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP026698 (20441..20543 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 89..94, 96..101 (AAATAA..TTATTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_unitig_2_pilon
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTTCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTTCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file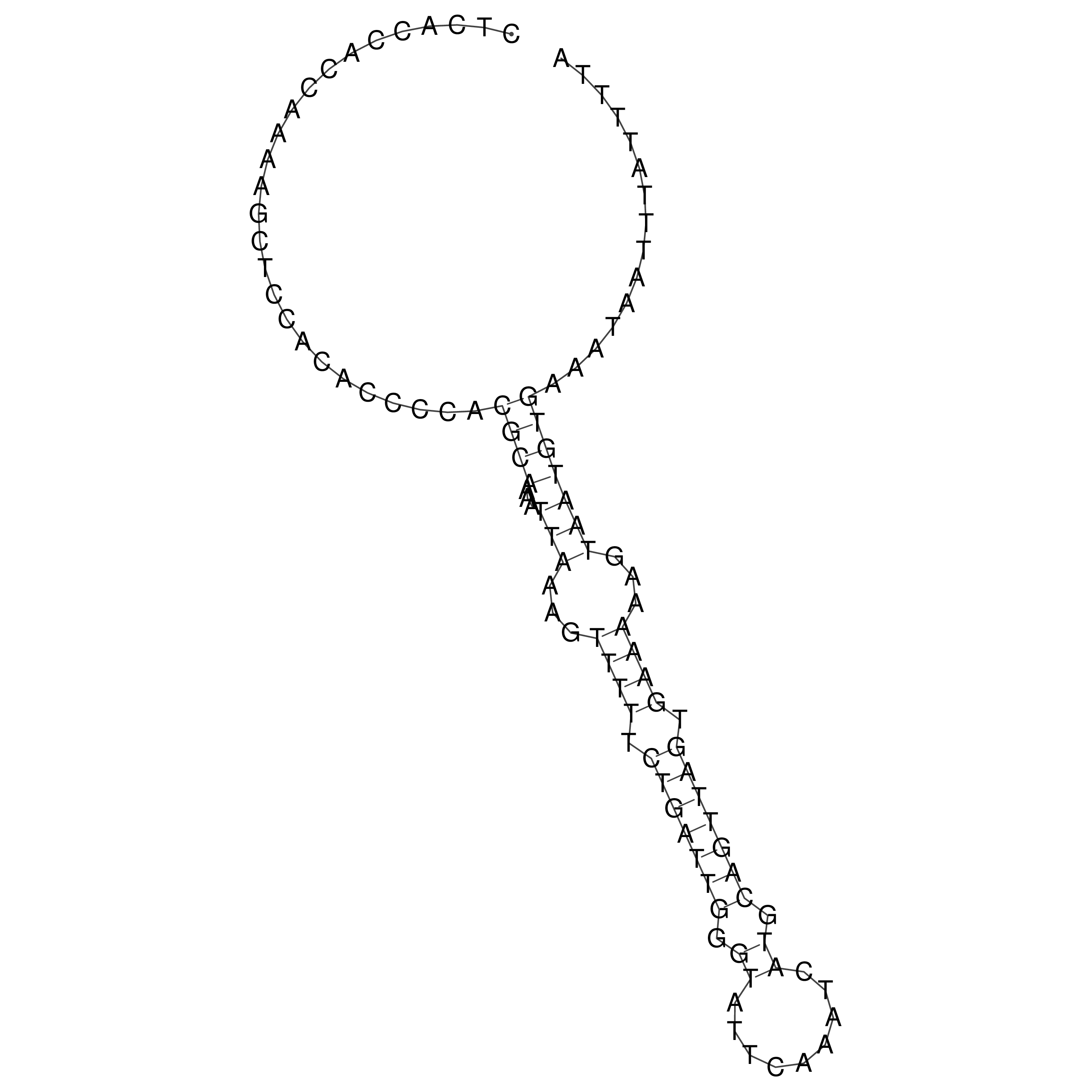
Relaxase
| ID | 11324 | GenBank | WP_022652257 |
| Name | traI_AM352_RS23845_unitig_2_pilon |
UniProt ID | A0A3S5I3M3 |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189292.49 Da Isoelectric Point: 6.1121
>WP_022652257.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVVPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVVPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13234 | GenBank | WP_008324134 |
| Name | traC_AM352_RS23760_unitig_2_pilon |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98841.51 Da Isoelectric Point: 5.7676
>WP_008324134.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacterales]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 13235 | GenBank | WP_023307203 |
| Name | traD_AM352_RS23840_unitig_2_pilon |
UniProt ID | _ |
| Length | 733 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 733 a.a. Molecular weight: 82212.80 Da Isoelectric Point: 5.1609
>WP_023307203.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQAE
QDELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQAE
QDELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 19893..46173
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AM352_RS23650 (AM352_23740) | 15077..15424 | - | 348 | WP_000609174 | IS66 family insertion sequence element accessory protein TnpB | - |
| AM352_RS23660 (AM352_23750) | 16361..16735 | + | 375 | WP_008324180 | hypothetical protein | - |
| AM352_RS23665 (AM352_23755) | 16789..17076 | + | 288 | WP_008324178 | hypothetical protein | - |
| AM352_RS23670 (AM352_23760) | 17123..17467 | + | 345 | WP_022652252 | hypothetical protein | - |
| AM352_RS23675 (AM352_23765) | 17553..17816 | + | 264 | WP_008324174 | hypothetical protein | - |
| AM352_RS23680 (AM352_23770) | 17859..18689 | + | 831 | WP_022652253 | N-6 DNA methylase | - |
| AM352_RS23685 (AM352_23775) | 19316..19846 | + | 531 | WP_008324170 | antirestriction protein | - |
| AM352_RS23690 (AM352_23780) | 19893..20369 | - | 477 | WP_008324168 | transglycosylase SLT domain-containing protein | virB1 |
| AM352_RS23695 (AM352_23785) | 20801..21193 | + | 393 | WP_008324167 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| AM352_RS23700 (AM352_23790) | 21382..22071 | + | 690 | WP_008324166 | hypothetical protein | - |
| AM352_RS23705 (AM352_23795) | 22158..22382 | + | 225 | WP_008324165 | TraY domain-containing protein | - |
| AM352_RS23710 (AM352_23800) | 22440..22790 | + | 351 | WP_008324162 | type IV conjugative transfer system pilin TraA | - |
| AM352_RS23715 (AM352_23805) | 22941..23246 | + | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| AM352_RS23720 (AM352_23810) | 23261..23827 | + | 567 | WP_008324158 | type IV conjugative transfer system protein TraE | traE |
| AM352_RS23725 (AM352_23815) | 23814..24557 | + | 744 | WP_008324157 | type-F conjugative transfer system secretin TraK | traK |
| AM352_RS23730 (AM352_23820) | 24544..25938 | + | 1395 | WP_008324156 | F-type conjugal transfer pilus assembly protein TraB | traB |
| AM352_RS23735 (AM352_23825) | 25960..26499 | + | 540 | WP_008324154 | type IV conjugative transfer system lipoprotein TraV | traV |
| AM352_RS23740 (AM352_23830) | 26619..26843 | + | 225 | WP_008324141 | TraR/DksA C4-type zinc finger protein | - |
| AM352_RS23745 (AM352_23835) | 26847..27032 | + | 186 | WP_022652254 | hypothetical protein | - |
| AM352_RS23750 (AM352_23840) | 27016..27417 | + | 402 | WP_008324137 | hypothetical protein | - |
| AM352_RS23755 (AM352_23845) | 27407..27973 | + | 567 | WP_008324135 | conjugal transfer pilus-stabilizing protein TraP | - |
| AM352_RS23760 (AM352_23850) | 27966..30596 | + | 2631 | WP_008324134 | type IV secretion system protein TraC | virb4 |
| AM352_RS23765 (AM352_23855) | 30593..30940 | + | 348 | WP_008324133 | type-F conjugative transfer system protein TrbI | - |
| AM352_RS23770 (AM352_23860) | 30940..31587 | + | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| AM352_RS23775 (AM352_23865) | 31590..32576 | + | 987 | WP_032160671 | conjugal transfer pilus assembly protein TraU | traU |
| AM352_RS23780 (AM352_23870) | 32593..33222 | + | 630 | WP_008324130 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| AM352_RS23785 (AM352_23875) | 33219..35072 | + | 1854 | WP_022652255 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| AM352_RS23790 (AM352_23880) | 35104..35292 | + | 189 | WP_008324128 | conjugal transfer protein TrbE | - |
| AM352_RS23795 (AM352_23885) | 35323..36072 | + | 750 | WP_008324127 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| AM352_RS23800 (AM352_23890) | 36089..36337 | + | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| AM352_RS23805 (AM352_23895) | 36327..36881 | + | 555 | WP_008324125 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| AM352_RS23810 (AM352_23900) | 36874..37302 | + | 429 | WP_008324122 | hypothetical protein | - |
| AM352_RS23815 (AM352_23905) | 37289..38659 | + | 1371 | WP_016247527 | conjugal transfer pilus assembly protein TraH | traH |
| AM352_RS23820 (AM352_23910) | 38659..41481 | + | 2823 | WP_008324119 | conjugal transfer mating-pair stabilization protein TraG | traG |
| AM352_RS23825 (AM352_23915) | 41503..41934 | + | 432 | WP_008324118 | entry exclusion protein | - |
| AM352_RS23830 (AM352_23920) | 42009..42683 | + | 675 | WP_008324909 | tetratricopeptide repeat protein | - |
| AM352_RS23835 (AM352_23925) | 42834..43865 | + | 1032 | WP_001395480 | IS630-like element ISEc33 family transposase | - |
| AM352_RS23840 (AM352_23930) | 43972..46173 | + | 2202 | WP_023307203 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 18256 | GenBank | NZ_CP026698 |
| Plasmid name | unitig_2_pilon | Incompatibility group | IncFII |
| Plasmid size | 82058 bp | Coordinate of oriT [Strand] | 20441..20543 [+] |
| Host baterium | Citrobacter koseri strain AR_0025 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |