Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117752 |
| Name | oriT_Res13-Abat-PEB01-P1-04-A|unnamednovel_1 |
| Organism | Raoultella terrigena strain Res13-Abat-PEB01-P1-04-A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP062917 (188820..188869 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_Res13-Abat-PEB01-P1-04-A|unnamednovel_1
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file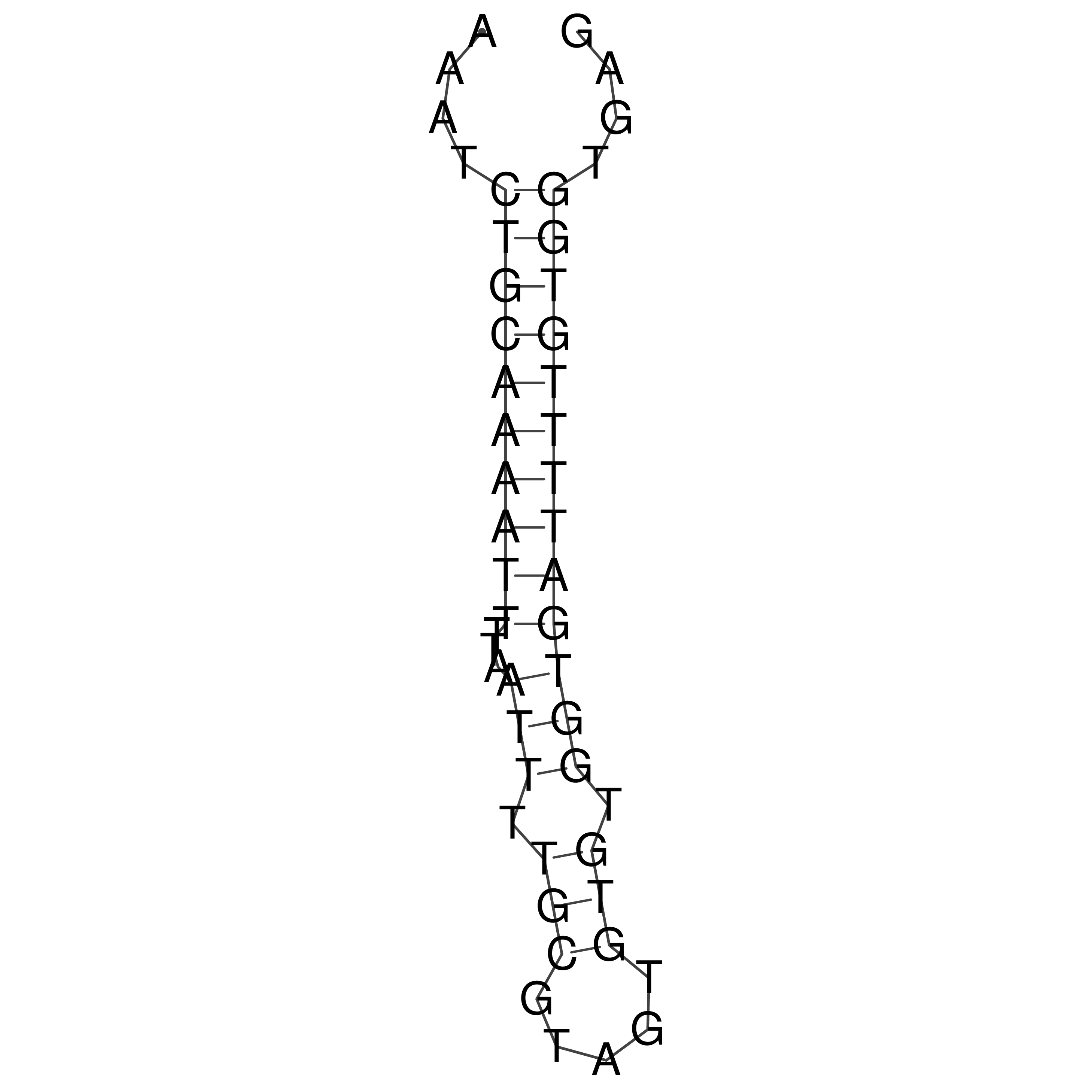
Relaxase
| ID | 11284 | GenBank | WP_195711719 |
| Name | traI_IMO34_RS27810_Res13-Abat-PEB01-P1-04-A|unnamednovel_1 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190160.51 Da Isoelectric Point: 5.7390
>WP_195711719.1 conjugative transfer relaxase/helicase TraI [Raoultella terrigena]
MLSFSQVKSAGSAGNYYTEKDNYYVTGSMEERWQGKGAEALGLEGKVDKQVFTDLLQGKIPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLATTRVQKDGVSETVLTGNLI
IARFNHDTSRAQEPQIHTHSVVINATRNGDKWQTLGSDTVGKTGFSENILANHIAFGKIYQNSLRADVES
MGYKTVDAGKHGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREDADKRATDIASAPVSPVKTDVPDISDVVTKTIAGLSDRKVQFTYADLLARTVSQLEA
KPGVFEQARNGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVKRQNHVSVTPDASVVREAPF
SDAVSVLAQDRPTMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETLSLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKENGVNTYR
WQGGKQATADIISEPEKGARYSRLAQEFAASVREGQESVAQISGSREQGILNGLIRETLKGEGVLGEKEL
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPENRQHDRFVIDRVTASSNMLTLRDKEGERLDMKVSAVD
SQWTLFRADKLPVAEGERLAVLGKIPETRLKGGEGVTVLKAEGGQLTVQRPGQKTTQTLAVGAGPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETELDAAIAQQKAGLHSPAEQAIHLSIPLLESNNLTFTRPQLLSTAMEFDGGGKVPMADIDRSI
QAQIKAGSLLSVPVAPGHGNDLLISRQTWDAEKSVLTRVLEGKAAMQPLMDRVPASLMTDLTSGQRAATR
MILETSDRFTVVQGYAGVGKTTQFRAVTGAIDLLPEETRPRVIGLGPTHRAVGEMQRAGVDAQTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDVNSALTTIEQVTPEQVPRKADAWVPENSVTQFTPAQEKAIV
DALKKGESVPADQPTTLYEALVMDYTGRTPEAQSQTLIITHLNNDRRALNSLIHDERVASGEVGKDEITL
PVLVTTNIRDGELRRLSTWAAHNGAVALVDNAYHRIGSVDKENQLITLTDSEGKERFISPREASAEGVTL
YRREDITVSQGDRMRFSKSDPERGYVANSVWEVKSVSGDSVTLSDGKTTRTLNPMEDEAQRHIDLAYAIT
AHGAQGASEPYAIALEGVAGGRKQMASFESTYVGLSRMKQHVQVYTDNREGWIKVIQASRSKATAHDILE
PRNDRAVKTADQLLSRARPLDESAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDSDGRLQVIGGEGRVMGNEDARFVALQGSRNGESLLAENMGEGVRVAREHPDSGVVVRLAGDDR
PWNPGAITGGSLWADKITQAPGADPVEIPLPPEVLAQKVADEAQRQEVEKQAERTAREVAGEDDKQRGPE
GQVKAVIDDVIRGVAGQKMQGERLTLPDDLEGREKAQAVQQVVSENLQRDRLQQMERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVTGSMEERWQGKGAEALGLEGKVDKQVFTDLLQGKIPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLATTRVQKDGVSETVLTGNLI
IARFNHDTSRAQEPQIHTHSVVINATRNGDKWQTLGSDTVGKTGFSENILANHIAFGKIYQNSLRADVES
MGYKTVDAGKHGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREDADKRATDIASAPVSPVKTDVPDISDVVTKTIAGLSDRKVQFTYADLLARTVSQLEA
KPGVFEQARNGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVKRQNHVSVTPDASVVREAPF
SDAVSVLAQDRPTMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETLSLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKENGVNTYR
WQGGKQATADIISEPEKGARYSRLAQEFAASVREGQESVAQISGSREQGILNGLIRETLKGEGVLGEKEL
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPENRQHDRFVIDRVTASSNMLTLRDKEGERLDMKVSAVD
SQWTLFRADKLPVAEGERLAVLGKIPETRLKGGEGVTVLKAEGGQLTVQRPGQKTTQTLAVGAGPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETELDAAIAQQKAGLHSPAEQAIHLSIPLLESNNLTFTRPQLLSTAMEFDGGGKVPMADIDRSI
QAQIKAGSLLSVPVAPGHGNDLLISRQTWDAEKSVLTRVLEGKAAMQPLMDRVPASLMTDLTSGQRAATR
MILETSDRFTVVQGYAGVGKTTQFRAVTGAIDLLPEETRPRVIGLGPTHRAVGEMQRAGVDAQTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDVNSALTTIEQVTPEQVPRKADAWVPENSVTQFTPAQEKAIV
DALKKGESVPADQPTTLYEALVMDYTGRTPEAQSQTLIITHLNNDRRALNSLIHDERVASGEVGKDEITL
PVLVTTNIRDGELRRLSTWAAHNGAVALVDNAYHRIGSVDKENQLITLTDSEGKERFISPREASAEGVTL
YRREDITVSQGDRMRFSKSDPERGYVANSVWEVKSVSGDSVTLSDGKTTRTLNPMEDEAQRHIDLAYAIT
AHGAQGASEPYAIALEGVAGGRKQMASFESTYVGLSRMKQHVQVYTDNREGWIKVIQASRSKATAHDILE
PRNDRAVKTADQLLSRARPLDESAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDSDGRLQVIGGEGRVMGNEDARFVALQGSRNGESLLAENMGEGVRVAREHPDSGVVVRLAGDDR
PWNPGAITGGSLWADKITQAPGADPVEIPLPPEVLAQKVADEAQRQEVEKQAERTAREVAGEDDKQRGPE
GQVKAVIDDVIRGVAGQKMQGERLTLPDDLEGREKAQAVQQVVSENLQRDRLQQMERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13173 | GenBank | WP_064349677 |
| Name | traD_IMO34_RS27815_Res13-Abat-PEB01-P1-04-A|unnamednovel_1 |
UniProt ID | _ |
| Length | 772 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 772 a.a. Molecular weight: 86728.66 Da Isoelectric Point: 4.9851
>WP_064349677.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella/Raoultella group]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGRALEYNSEQILADKYTIWCGEQLWTSFVFAAIVSLTVCIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKYSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDARCAAWDIWSECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFSEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLKNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLGMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKSRPKIAEGFILRNLDTRTDARLDTLLEAREAESSHARMLFTPDAPEPEGDDKDRTASEQHEPVKQPE
PTGDPVPPVPVKAPTAAKTSVATEPVKSPETPQSVIPATRVTTVPLIRPKPASAATGTAAVAGATAHTGG
TEQELAQQAADSGQEMMPAGMNEDGEIEDMQAYDDWLAGEQTQRDMQRREEVNINNSHRRDEQDDIEIGG
NF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGRALEYNSEQILADKYTIWCGEQLWTSFVFAAIVSLTVCIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKYSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDARCAAWDIWSECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFSEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLKNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLGMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKSRPKIAEGFILRNLDTRTDARLDTLLEAREAESSHARMLFTPDAPEPEGDDKDRTASEQHEPVKQPE
PTGDPVPPVPVKAPTAAKTSVATEPVKSPETPQSVIPATRVTTVPLIRPKPASAATGTAAVAGATAHTGG
TEQELAQQAADSGQEMMPAGMNEDGEIEDMQAYDDWLAGEQTQRDMQRREEVNINNSHRRDEQDDIEIGG
NF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 160642..186441
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IMO34_RS27815 (IMO34_27810) | 160642..162960 | - | 2319 | WP_064349677 | type IV conjugative transfer system coupling protein TraD | virb4 |
| IMO34_RS27820 (IMO34_27815) | 163155..163886 | - | 732 | WP_064349680 | conjugal transfer complement resistance protein TraT | - |
| IMO34_RS27825 (IMO34_27820) | 164080..164607 | - | 528 | WP_195711720 | conjugal transfer protein TraS | - |
| IMO34_RS27830 (IMO34_27825) | 164613..167468 | - | 2856 | WP_195711721 | conjugal transfer mating-pair stabilization protein TraG | traG |
| IMO34_RS27835 (IMO34_27830) | 167468..168838 | - | 1371 | WP_195711722 | conjugal transfer pilus assembly protein TraH | traH |
| IMO34_RS27840 (IMO34_27835) | 168825..169253 | - | 429 | WP_101856563 | conjugal transfer protein TrbF | - |
| IMO34_RS27845 (IMO34_27840) | 169246..169812 | - | 567 | WP_101856562 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| IMO34_RS27850 (IMO34_27845) | 169787..170023 | - | 237 | WP_101856561 | type-F conjugative transfer system pilin chaperone TraQ | - |
| IMO34_RS27855 (IMO34_27850) | 170034..170786 | - | 753 | WP_064361002 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| IMO34_RS27860 (IMO34_27855) | 170809..171135 | - | 327 | WP_101856560 | hypothetical protein | - |
| IMO34_RS27865 (IMO34_27860) | 171361..171597 | - | 237 | WP_101856558 | conjugal transfer protein TrbE | - |
| IMO34_RS27870 (IMO34_27865) | 171587..171967 | - | 381 | WP_064360999 | hypothetical protein | - |
| IMO34_RS27875 (IMO34_27870) | 172121..172591 | - | 471 | WP_075809048 | hypothetical protein | - |
| IMO34_RS27880 (IMO34_27875) | 172675..174630 | - | 1956 | WP_064349681 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| IMO34_RS27885 (IMO34_27880) | 174630..175259 | - | 630 | WP_064349684 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| IMO34_RS27890 (IMO34_27885) | 175272..176261 | - | 990 | WP_064349687 | conjugal transfer pilus assembly protein TraU | traU |
| IMO34_RS27895 (IMO34_27890) | 176272..176901 | - | 630 | WP_195711723 | type-F conjugative transfer system protein TraW | traW |
| IMO34_RS27900 (IMO34_27895) | 176898..177290 | - | 393 | WP_064349691 | type-F conjugative transfer system protein TrbI | - |
| IMO34_RS27905 (IMO34_27900) | 177290..179929 | - | 2640 | WP_064349692 | type IV secretion system protein TraC | virb4 |
| IMO34_RS27910 (IMO34_27905) | 180014..180394 | - | 381 | WP_064349750 | hypothetical protein | - |
| IMO34_RS27915 (IMO34_27910) | 180409..180756 | - | 348 | WP_074180877 | hypothetical protein | - |
| IMO34_RS27920 (IMO34_27915) | 180753..181157 | - | 405 | WP_064349693 | hypothetical protein | - |
| IMO34_RS27925 (IMO34_27920) | 181154..181375 | - | 222 | WP_064349696 | hypothetical protein | - |
| IMO34_RS27930 (IMO34_27925) | 181400..181615 | - | 216 | WP_064349698 | hypothetical protein | - |
| IMO34_RS27935 (IMO34_27930) | 182226..182795 | - | 570 | WP_195711726 | type IV conjugative transfer system lipoprotein TraV | traV |
| IMO34_RS27940 (IMO34_27935) | 182818..183414 | - | 597 | WP_064349701 | conjugal transfer pilus-stabilizing protein TraP | - |
| IMO34_RS27945 (IMO34_27940) | 183407..184828 | - | 1422 | WP_064349704 | F-type conjugal transfer pilus assembly protein TraB | traB |
| IMO34_RS27950 (IMO34_27945) | 184828..185565 | - | 738 | WP_064349705 | type-F conjugative transfer system secretin TraK | traK |
| IMO34_RS27955 (IMO34_27950) | 185552..186118 | - | 567 | WP_064349707 | type IV conjugative transfer system protein TraE | traE |
| IMO34_RS27960 (IMO34_27955) | 186136..186441 | - | 306 | WP_064349709 | type IV conjugative transfer system protein TraL | traL |
| IMO34_RS27965 (IMO34_27960) | 186455..186823 | - | 369 | WP_064349711 | type IV conjugative transfer system pilin TraA | - |
| IMO34_RS27970 (IMO34_27965) | 186892..187092 | - | 201 | WP_074180879 | TraY domain-containing protein | - |
| IMO34_RS27975 (IMO34_27970) | 187178..187882 | - | 705 | WP_064349713 | hypothetical protein | - |
| IMO34_RS27980 (IMO34_27975) | 188119..188511 | - | 393 | WP_064349715 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| IMO34_RS27985 (IMO34_27980) | 188944..189420 | + | 477 | WP_064349718 | transglycosylase SLT domain-containing protein | - |
| IMO34_RS27990 (IMO34_27985) | 189458..189987 | - | 530 | Protein_192 | antirestriction protein | - |
| IMO34_RS27995 (IMO34_27990) | 190805..190951 | - | 147 | WP_064349719 | hypothetical protein | - |
| IMO34_RS28000 (IMO34_27995) | 191045..191392 | - | 348 | WP_064349720 | hypothetical protein | - |
Host bacterium
| ID | 18185 | GenBank | NZ_CP062917 |
| Plasmid name | Res13-Abat-PEB01-P1-04-A|unnamednovel_1 | Incompatibility group | IncFIB |
| Plasmid size | 218566 bp | Coordinate of oriT [Strand] | 188820..188869 [+] |
| Host baterium | Raoultella terrigena strain Res13-Abat-PEB01-P1-04-A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsC, arsB, arsR, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |