Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117709 |
| Name | oriT_p117885-NR |
| Organism | Raoultella ornithinolytica strain 172117885 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MT074086 (34448..34497 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..13, 18..24 (GCAAAAT..ATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_p117885-NR
AAATCTGCAAAATATTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATATTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file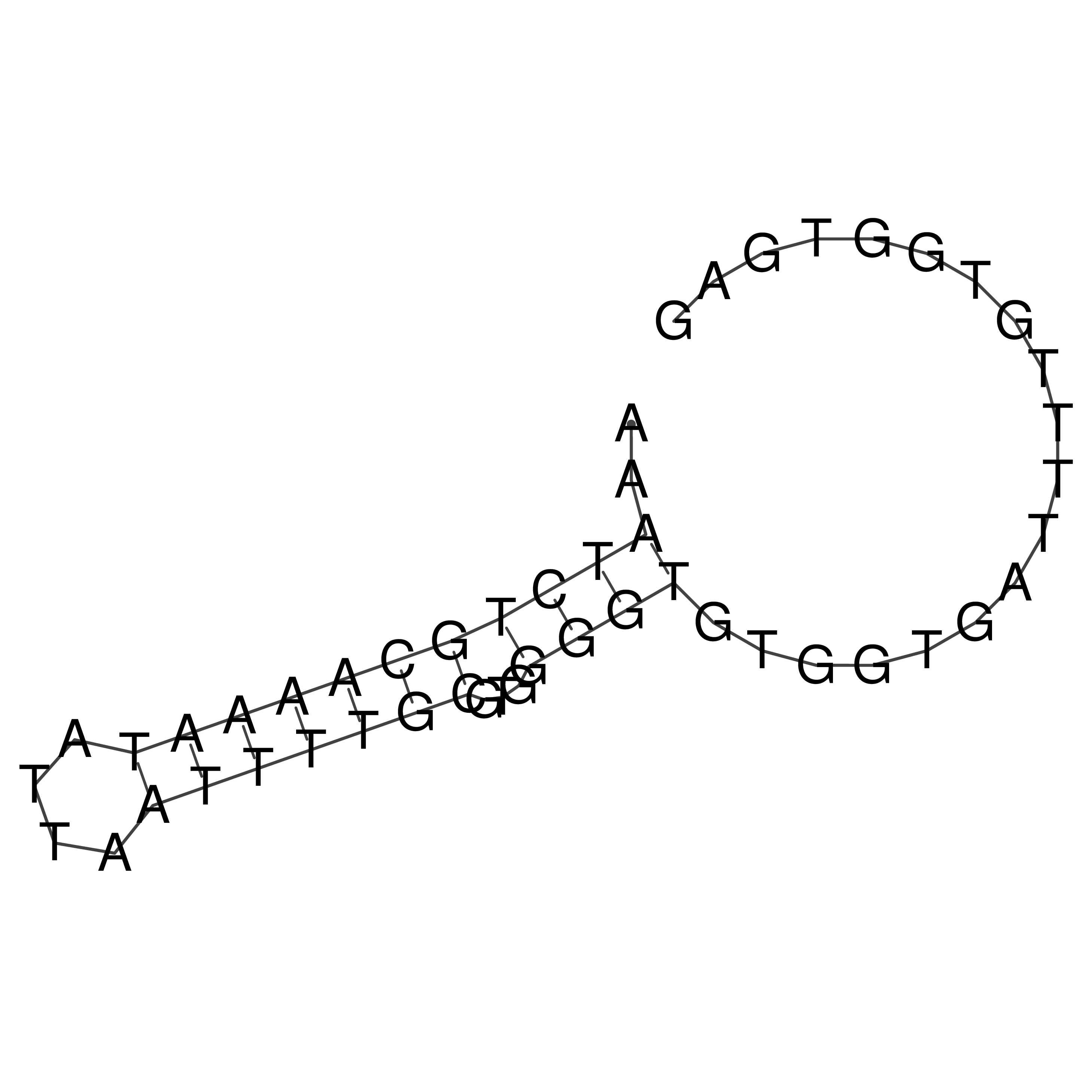
Relaxase
| ID | 11264 | GenBank | WP_076752116 |
| Name | traI_I0Q10_RS00375_p117885-NR |
UniProt ID | A0A9Q9JMY5 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190488.11 Da Isoelectric Point: 5.7342
>WP_076752116.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella/Raoultella group]
MMSIGSVKSAGSASNYYTDKDNYYVIGSMDERWQGKGAEALGLDGKVDKQVFTDLLKGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSVMAMIGGDKRLIDAHNQAVTEAVKQVETLAATRVMTDGKSETVLTGNLI
VAKFNHDTNRNQEPQLHTHAVVINATQNGGKWQSLGTDTVGKTGFIENVYANQIAFGKLYREVLKPLVEK
LGYETEVTGKHGMWEMKDVPVEPFSTRSQEVREAAGPDASLKSRDVATLDTRKSKETLDPAEKMVEWVNT
LKESGFDIHSYREDADKRAAEINRAPVSPAKTDGPDISDVVTKAIAGLSDRKVQFTYADMLARTVSQLEA
KPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRHAPF
SDAVSVLAQDRPAMGIVYGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLVGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETLSLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGNQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGVREQGVLNGIIREVLKNEGTLGAKEM
TVTTLTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFLIDRVTASSNMLTLRDKEGERLDLKVSAVD
SQWSLFRADKLPVASGERLSVLGKIPETRLKGGESITVLNAEAGQLTVQRPGQKTTQTLAVGAGPFDGIK
VGHGWVESPGRSVSETATVFASLTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIALQKAGLHTPAEQVIHLSIPLLESEELTFTRPQLLATALETGGGKVPMTEIDATIQ
AQIKAGSLLNVPVAPGHGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDSVPGSLMTDLTSGQRASTRM
ILETTDRFTVVQGYAGVGKTTQLRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVNRALATIEQITPEQVPRKDGAWVPENSVVEFTPAQEETIQK
ALNKGESVPAGQPTTLYEALVKDYADRTPEAQSQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKTTRTLNPKADQAQQHVDLAYAITA
HGAQGASEPFAIALEGVAGGRERMASFESSYVGLSRMKQHVQVYTDNREGWIRAINHSPEKATAHDILEP
RNDRAVKTAGQLFSRARALDEAAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDNDGRLQVIGGEGRVMGNEEARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMERQTEQATREVSGEEKKVGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVATESLQRDRLQQMERDMVRDLSREKTLG
GD
MMSIGSVKSAGSASNYYTDKDNYYVIGSMDERWQGKGAEALGLDGKVDKQVFTDLLKGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSVMAMIGGDKRLIDAHNQAVTEAVKQVETLAATRVMTDGKSETVLTGNLI
VAKFNHDTNRNQEPQLHTHAVVINATQNGGKWQSLGTDTVGKTGFIENVYANQIAFGKLYREVLKPLVEK
LGYETEVTGKHGMWEMKDVPVEPFSTRSQEVREAAGPDASLKSRDVATLDTRKSKETLDPAEKMVEWVNT
LKESGFDIHSYREDADKRAAEINRAPVSPAKTDGPDISDVVTKAIAGLSDRKVQFTYADMLARTVSQLEA
KPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRHAPF
SDAVSVLAQDRPAMGIVYGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLVGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETLSLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGNQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGVREQGVLNGIIREVLKNEGTLGAKEM
TVTTLTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFLIDRVTASSNMLTLRDKEGERLDLKVSAVD
SQWSLFRADKLPVASGERLSVLGKIPETRLKGGESITVLNAEAGQLTVQRPGQKTTQTLAVGAGPFDGIK
VGHGWVESPGRSVSETATVFASLTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIALQKAGLHTPAEQVIHLSIPLLESEELTFTRPQLLATALETGGGKVPMTEIDATIQ
AQIKAGSLLNVPVAPGHGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDSVPGSLMTDLTSGQRASTRM
ILETTDRFTVVQGYAGVGKTTQLRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVNRALATIEQITPEQVPRKDGAWVPENSVVEFTPAQEETIQK
ALNKGESVPAGQPTTLYEALVKDYADRTPEAQSQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKTTRTLNPKADQAQQHVDLAYAITA
HGAQGASEPFAIALEGVAGGRERMASFESSYVGLSRMKQHVQVYTDNREGWIRAINHSPEKATAHDILEP
RNDRAVKTAGQLFSRARALDEAAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDNDGRLQVIGGEGRVMGNEEARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMERQTEQATREVSGEEKKVGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVATESLQRDRLQQMERDMVRDLSREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13151 | GenBank | WP_032736842 |
| Name | traD_I0Q10_RS00370_p117885-NR |
UniProt ID | _ |
| Length | 764 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 764 a.a. Molecular weight: 85398.61 Da Isoelectric Point: 5.0675
>WP_032736842.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTISYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTICVVTFFVASWVLGRQGKL
QSEDENTGGRQLSDKPKDVARQMKRDGAASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMRGDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYNPRPKIAEGFIQRRLDTRIDERLSAQLEAREAEGSLVRALFTPDEPVSEPAEEKAGVQPEQTKPAKDS
VLPVPVKTSSMAAEGAGKTPAVPLSGAPVTRVKTVPLIRPKPAASVTGTAAAASTAAVSATAGGTEQELA
QQPVEADQDMLPAGMNEDGVIEDMQAYDAWLEGEQIQRDMQRREEVNINHSRSEQDDIEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTISYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTICVVTFFVASWVLGRQGKL
QSEDENTGGRQLSDKPKDVARQMKRDGAASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMRGDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYNPRPKIAEGFIQRRLDTRIDERLSAQLEAREAEGSLVRALFTPDEPVSEPAEEKAGVQPEQTKPAKDS
VLPVPVKTSSMAAEGAGKTPAVPLSGAPVTRVKTVPLIRPKPAASVTGTAAAASTAAVSATAGGTEQELA
QQPVEADQDMLPAGMNEDGVIEDMQAYDAWLEGEQIQRDMQRREEVNINHSRSEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 36873..59933
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| I0Q10_RS00205 | 32907..33137 | - | 231 | WP_076752135 | hypothetical protein | - |
| I0Q10_RS00210 | 33329..33859 | + | 531 | WP_076752134 | antirestriction protein | - |
| I0Q10_RS00215 | 33897..34373 | - | 477 | WP_076752132 | transglycosylase SLT domain-containing protein | - |
| I0Q10_RS00220 | 34805..35197 | + | 393 | WP_032736874 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| I0Q10_RS00225 | 35445..36137 | + | 693 | WP_181717165 | hypothetical protein | - |
| I0Q10_RS00230 | 36222..36422 | + | 201 | WP_060415469 | TraY domain-containing protein | - |
| I0Q10_RS00235 | 36491..36859 | + | 369 | WP_032736871 | type IV conjugative transfer system pilin TraA | - |
| I0Q10_RS00240 | 36873..37178 | + | 306 | WP_032736870 | type IV conjugative transfer system protein TraL | traL |
| I0Q10_RS00245 | 37198..37764 | + | 567 | WP_032736869 | type IV conjugative transfer system protein TraE | traE |
| I0Q10_RS00250 | 37751..38485 | + | 735 | WP_032736867 | type-F conjugative transfer system secretin TraK | traK |
| I0Q10_RS00255 | 38485..39906 | + | 1422 | WP_032736866 | F-type conjugal transfer pilus assembly protein TraB | traB |
| I0Q10_RS00260 | 39899..40060 | + | 162 | WP_154235504 | hypothetical protein | - |
| I0Q10_RS00265 | 40080..40649 | + | 570 | WP_042946293 | type IV conjugative transfer system lipoprotein TraV | traV |
| I0Q10_RS00270 | 40761..41039 | + | 279 | WP_032736865 | hypothetical protein | - |
| I0Q10_RS00990 | 41124..41525 | + | 402 | WP_032736864 | hypothetical protein | - |
| I0Q10_RS00280 | 41522..41821 | + | 300 | WP_032736863 | hypothetical protein | - |
| I0Q10_RS00285 | 41909..42292 | + | 384 | WP_042946290 | hypothetical protein | - |
| I0Q10_RS00290 | 42385..45024 | + | 2640 | WP_032736861 | type IV secretion system protein TraC | virb4 |
| I0Q10_RS00295 | 45024..45413 | + | 390 | WP_032736860 | type-F conjugative transfer system protein TrbI | - |
| I0Q10_RS00300 | 45410..46036 | + | 627 | WP_042946288 | type-F conjugative transfer system protein TraW | traW |
| I0Q10_RS00305 | 46080..47039 | + | 960 | WP_088912317 | conjugal transfer pilus assembly protein TraU | traU |
| I0Q10_RS00310 | 47052..47678 | + | 627 | WP_032736857 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| I0Q10_RS00315 | 47675..49618 | + | 1944 | WP_032736856 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| I0Q10_RS00320 | 49780..50343 | + | 564 | WP_195156997 | hypothetical protein | - |
| I0Q10_RS00325 | 50333..50569 | + | 237 | WP_076752122 | conjugal transfer protein TrbE | - |
| I0Q10_RS00330 | 50566..50751 | + | 186 | WP_032736852 | hypothetical protein | - |
| I0Q10_RS00335 | 50798..51124 | + | 327 | WP_032736851 | hypothetical protein | - |
| I0Q10_RS00340 | 51145..51897 | + | 753 | WP_042946285 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| I0Q10_RS00345 | 51908..52144 | + | 237 | WP_032736848 | type-F conjugative transfer system pilin chaperone TraQ | - |
| I0Q10_RS00350 | 52131..52694 | + | 564 | WP_224250969 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| I0Q10_RS00355 | 52681..54051 | + | 1371 | WP_076752120 | conjugal transfer pilus assembly protein TraH | traH |
| I0Q10_RS00360 | 54051..56894 | + | 2844 | WP_076752118 | conjugal transfer mating-pair stabilization protein TraG | traG |
| I0Q10_RS00365 | 56905..57432 | + | 528 | WP_032736843 | hypothetical protein | - |
| I0Q10_RS00370 | 57639..59933 | + | 2295 | WP_032736842 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 18142 | GenBank | NZ_MT074086 |
| Plasmid name | p117885-NR | Incompatibility group | IncFII |
| Plasmid size | 184801 bp | Coordinate of oriT [Strand] | 34448..34497 [-] |
| Host baterium | Raoultella ornithinolytica strain 172117885 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsR, arsD, arsA, arsB, arsC, arsH, silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |