Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117660 |
| Name | oriT_NDM-101|unnamed1 |
| Organism | Klebsiella quasipneumoniae strain NDM-101 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP085198 (51769..51818 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_NDM-101|unnamed1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file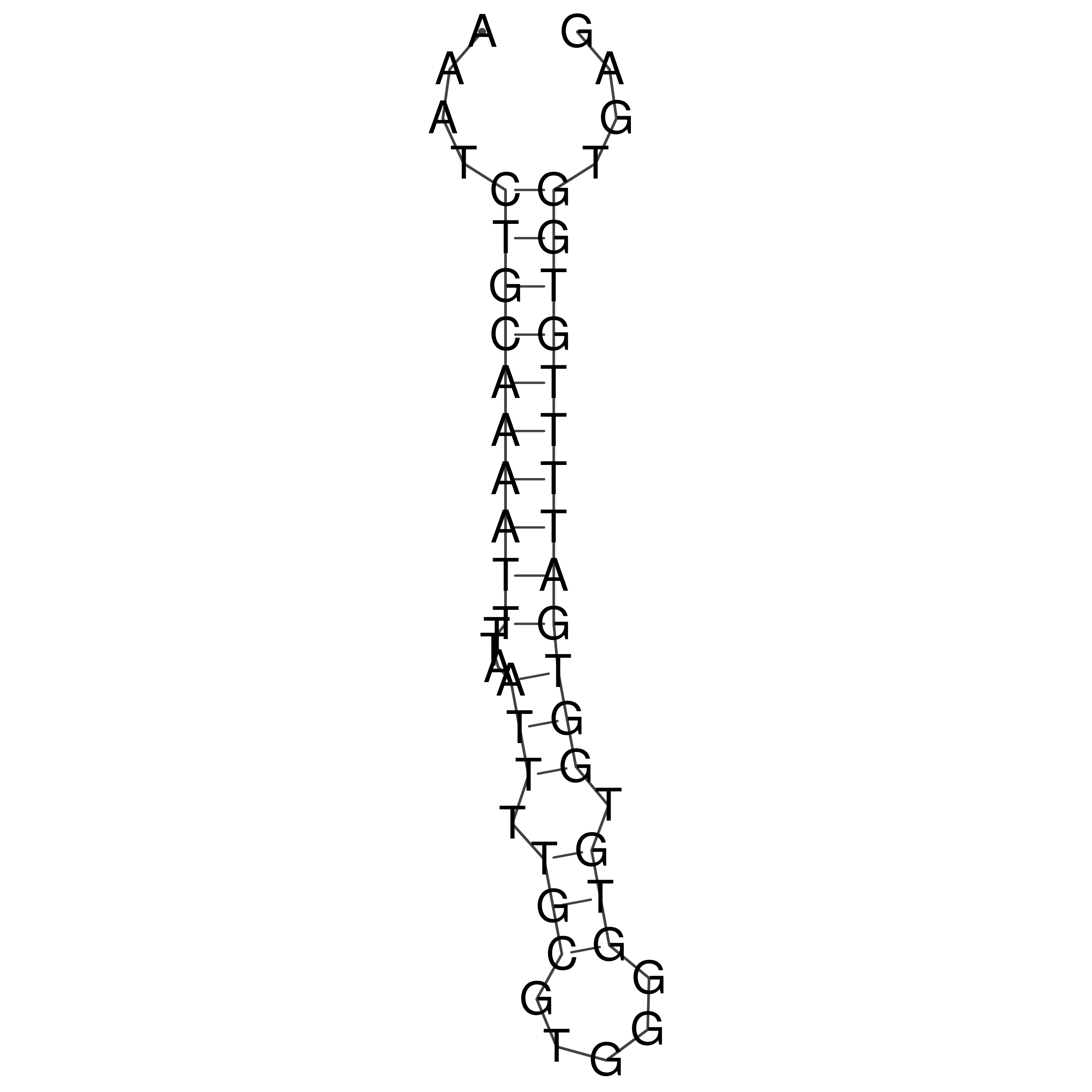
Relaxase
| ID | 11240 | GenBank | WP_265049843 |
| Name | traI_LH783_RS25570_NDM-101|unnamed1 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190626.68 Da Isoelectric Point: 6.1561
>WP_265049843.1 conjugative transfer relaxase/helicase TraI [Klebsiella quasipneumoniae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKEVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLVGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVLGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKEVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLVGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVLGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 51217..66068
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LH783_RS25400 (LH783_25435) | 46426..46752 | + | 327 | WP_038992719 | hypothetical protein | - |
| LH783_RS25405 (LH783_25440) | 46911..47063 | + | 153 | WP_224230667 | DUF5431 family protein | - |
| LH783_RS25410 (LH783_25445) | 47008..47127 | + | 120 | WP_223156217 | type I toxin-antitoxin system Hok family toxin | - |
| LH783_RS25415 (LH783_25450) | 48032..48412 | + | 381 | WP_038990973 | hypothetical protein | - |
| LH783_RS25420 (LH783_25455) | 48479..48826 | + | 348 | WP_038990975 | hypothetical protein | - |
| LH783_RS25425 (LH783_25460) | 48914..49144 | + | 231 | WP_038990977 | hypothetical protein | - |
| LH783_RS25430 (LH783_25465) | 49191..50024 | + | 834 | WP_038990979 | N-6 DNA methylase | - |
| LH783_RS25435 (LH783_25470) | 50649..51179 | + | 531 | WP_023292160 | antirestriction protein | - |
| LH783_RS25440 (LH783_25475) | 51217..51624 | - | 408 | WP_225375048 | transglycosylase SLT domain-containing protein | virB1 |
| LH783_RS25445 (LH783_25480) | 52130..52522 | + | 393 | WP_004178062 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LH783_RS25450 (LH783_25485) | 52729..53424 | + | 696 | WP_023292158 | transcriptional regulator TraJ family protein | - |
| LH783_RS25455 (LH783_25490) | 53508..53879 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| LH783_RS25460 (LH783_25495) | 53933..54301 | + | 369 | WP_004178060 | type IV conjugative transfer system pilin TraA | - |
| LH783_RS25465 (LH783_25500) | 54315..54620 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| LH783_RS25470 (LH783_25505) | 54640..55206 | + | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| LH783_RS25475 (LH783_25510) | 55193..55933 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| LH783_RS25480 (LH783_25515) | 55933..57357 | + | 1425 | WP_023292156 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LH783_RS25485 (LH783_25520) | 57354..57539 | + | 186 | WP_038990989 | hypothetical protein | - |
| LH783_RS25490 (LH783_25525) | 57558..58127 | + | 570 | WP_032429300 | type IV conjugative transfer system lipoprotein TraV | traV |
| LH783_RS25495 (LH783_25530) | 58259..58669 | + | 411 | WP_023292154 | hypothetical protein | - |
| LH783_RS25500 (LH783_25535) | 58674..58964 | + | 291 | WP_032429299 | hypothetical protein | - |
| LH783_RS25505 (LH783_25540) | 58988..59206 | + | 219 | WP_004171484 | hypothetical protein | - |
| LH783_RS25510 (LH783_25545) | 59207..59524 | + | 318 | WP_023292153 | hypothetical protein | - |
| LH783_RS25515 (LH783_25550) | 59591..59995 | + | 405 | WP_023292152 | hypothetical protein | - |
| LH783_RS25520 (LH783_25555) | 59992..60282 | + | 291 | WP_023292151 | hypothetical protein | - |
| LH783_RS25525 (LH783_25560) | 60290..60688 | + | 399 | WP_004153071 | hypothetical protein | - |
| LH783_RS25530 (LH783_25565) | 60760..63399 | + | 2640 | WP_004152592 | type IV secretion system protein TraC | virb4 |
| LH783_RS25535 (LH783_25570) | 63399..63788 | + | 390 | WP_004152591 | type-F conjugative transfer system protein TrbI | - |
| LH783_RS25540 (LH783_25575) | 63788..64414 | + | 627 | WP_004152590 | type-F conjugative transfer system protein TraW | traW |
| LH783_RS25545 (LH783_25580) | 64458..65417 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| LH783_RS25550 (LH783_25585) | 65430..66068 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LH783_RS25555 (LH783_25590) | 66116..67765 | + | 1650 | Protein_85 | conjugal transfer protein TraN | - |
| LH783_RS27700 | 67743..67931 | - | 189 | WP_320109480 | hypothetical protein | - |
| LH783_RS25560 (LH783_25595) | 67913..68893 | - | 981 | WP_000019445 | IS5-like element ISKpn26 family transposase | - |
Host bacterium
| ID | 18093 | GenBank | NZ_CP085198 |
| Plasmid name | NDM-101|unnamed1 | Incompatibility group | IncFIB |
| Plasmid size | 183890 bp | Coordinate of oriT [Strand] | 51769..51818 [-] |
| Host baterium | Klebsiella quasipneumoniae strain NDM-101 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | fecE, fecD, pcoE, pcoS, pcoR, pcoD, pcoC, pcoB, pcoA, silP, silA, silB, silF, silC, silR, silS, silE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |