Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117594 |
| Name | oriT_pWP3-S18-ESBL-03_4 |
| Organism | Klebsiella quasipneumoniae strain WP3-S18-ESBL-03 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP022018 (70277..70325 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pWP3-S18-ESBL-03_4
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file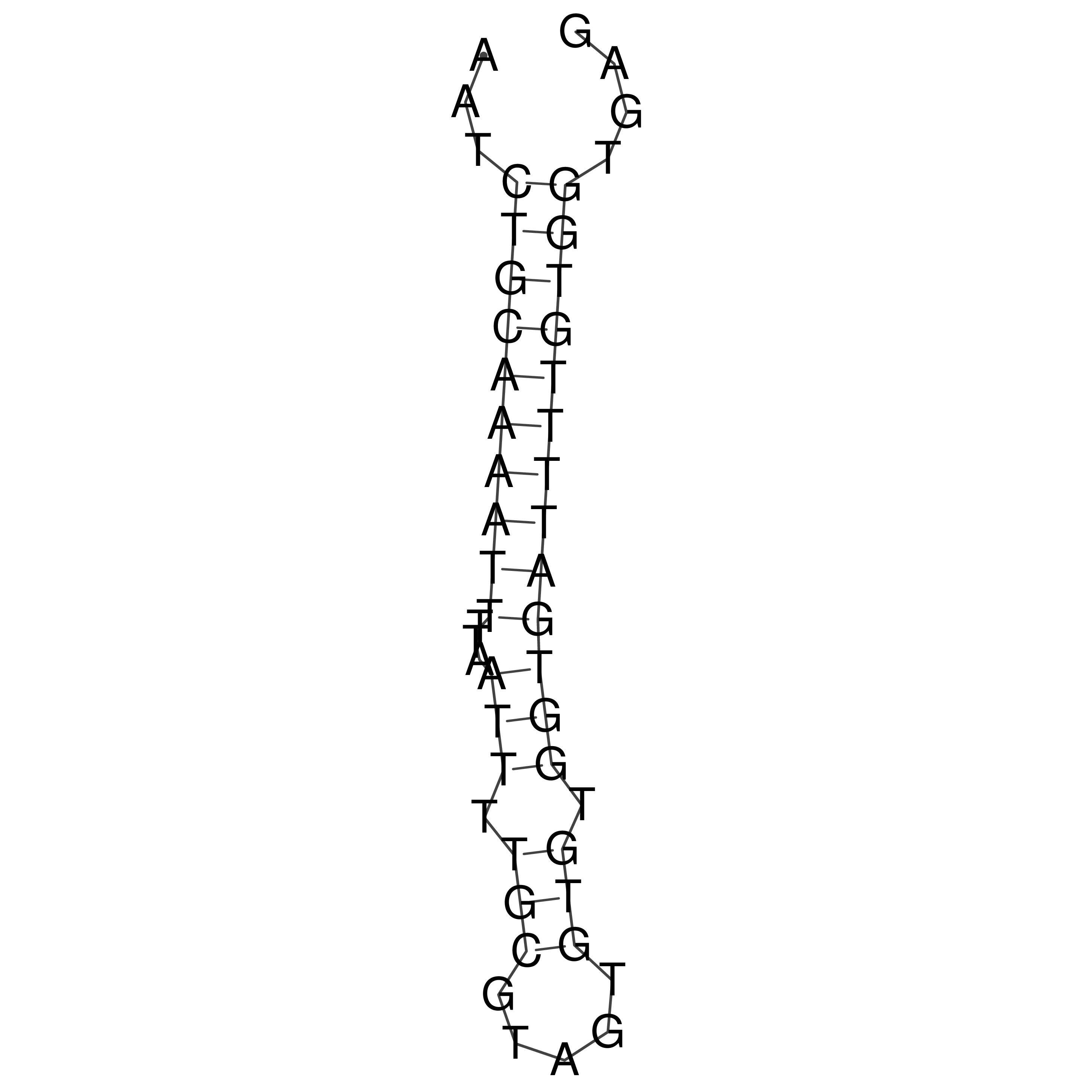
Relaxase
| ID | 11215 | GenBank | WP_182972118 |
| Name | traI_H7R87_RS29775_pWP3-S18-ESBL-03_4 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190096.40 Da Isoelectric Point: 5.9146
>WP_182972118.1 conjugative transfer relaxase/helicase TraI [Klebsiella quasipneumoniae]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDASVVREAPF
SDAVSVLAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGKQATADIISEPDKSARYSRLAQEFAASVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGNRDM
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDREGERLDLKVSAVD
SQWTLFRTDKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLPVGSSPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETDLEEAIAQQKAGLHTPAEQAIHLSVPLLESNSLTFTRPQLLATALETGGGKVPMKDIEATIQ
AQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMGRVPGSLMTDLTAGQRAATRM
ILETPDRFTVVQGYAGVGKTTQFKAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLQQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVSRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQD
ALKEGKEIPPDTPASLHDAIVKDYTGRTPEAQANTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTLP
VLVTSNIRDGELRKLSTWSEHQGAVALLDNIYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTLY
RPEHITVSQGDRMRFSKSDPERGYVANSVWEVKAVSGDSVTLSGGKTTRTLNPKADEAQRHVDLAYAITA
HGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLEP
QNARAVKNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGIW
LSPLTDRDGRLEAIGGEGRIMGDETARFVALQNSRNGESLLAGNMGEGVRTARDHPDSGVIVRLAGDDRP
WNPEAITGGRIWADPLPNPPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQATREVAGEEKKAGEPG
ERIKEVIGDVIRGLERDRPGEEKTLLPDDPQTRRQEAAIQQVASESLQRERLQQMERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDASVVREAPF
SDAVSVLAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGKQATADIISEPDKSARYSRLAQEFAASVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGNRDM
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDREGERLDLKVSAVD
SQWTLFRTDKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLPVGSSPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDAARTTEKLARHTAFSVVSEQ
LKARSGETDLEEAIAQQKAGLHTPAEQAIHLSVPLLESNSLTFTRPQLLATALETGGGKVPMKDIEATIQ
AQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMGRVPGSLMTDLTAGQRAATRM
ILETPDRFTVVQGYAGVGKTTQFKAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLQQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVSRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQD
ALKEGKEIPPDTPASLHDAIVKDYTGRTPEAQANTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTLP
VLVTSNIRDGELRKLSTWSEHQGAVALLDNIYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTLY
RPEHITVSQGDRMRFSKSDPERGYVANSVWEVKAVSGDSVTLSGGKTTRTLNPKADEAQRHVDLAYAITA
HGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLEP
QNARAVKNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGIW
LSPLTDRDGRLEAIGGEGRIMGDETARFVALQNSRNGESLLAGNMGEGVRTARDHPDSGVIVRLAGDDRP
WNPEAITGGRIWADPLPNPPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQATREVAGEEKKAGEPG
ERIKEVIGDVIRGLERDRPGEEKTLLPDDPQTRRQEAAIQQVASESLQRERLQQMERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6484 | GenBank | WP_032426796 |
| Name | WP_032426796_pWP3-S18-ESBL-03_4 |
UniProt ID | _ |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15036.10 Da Isoelectric Point: 4.5326
>WP_032426796.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacterales]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 13099 | GenBank | WP_020803851 |
| Name | traD_H7R87_RS29770_pWP3-S18-ESBL-03_4 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85873.87 Da Isoelectric Point: 4.9596
>WP_020803851.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFIFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAM
KYKPRPKIAEGFIQRTPDTRADARLSALLEAREAEGSLARALFTPDAPELAGKDSTPGEQPEPVSQPAPA
EVAVSPAPVNAPPTTKSPAAEPSAGAPASPPPEATVLRNTTVPLIKPKAAGTAATASSAGTPVAPAGGTE
QELVQHSTEQGRDMLPAGMNEDGVIEDMQAYDAWLADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFIFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAM
KYKPRPKIAEGFIQRTPDTRADARLSALLEAREAEGSLARALFTPDAPELAGKDSTPGEQPEPVSQPAPA
EVAVSPAPVNAPPTTKSPAAEPSAGAPASPPPEATVLRNTTVPLIKPKAAGTAATASSAGTPVAPAGGTE
QELVQHSTEQGRDMLPAGMNEDGVIEDMQAYDAWLADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 72744..98842
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H7R87_RS29575 (WP3S18E03_P40820) | 67953..68183 | + | 231 | WP_020314756 | hypothetical protein | - |
| H7R87_RS29580 (WP3S18E03_P40830) | 68281..68496 | + | 216 | Protein_91 | SAM-dependent DNA methyltransferase | - |
| H7R87_RS29585 (WP3S18E03_P40840) | 69145..69687 | + | 543 | WP_032426797 | antirestriction protein | - |
| H7R87_RS29590 (WP3S18E03_P40850) | 69710..70132 | - | 423 | WP_020315406 | transglycosylase SLT domain-containing protein | - |
| H7R87_RS29595 (WP3S18E03_P40860) | 70638..71030 | + | 393 | WP_032426796 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| H7R87_RS29600 (WP3S18E03_P40870) | 71268..71960 | + | 693 | WP_032426795 | conjugal transfer protein TrbJ | - |
| H7R87_RS29605 | 72136..72300 | + | 165 | WP_032444506 | TraY domain-containing protein | - |
| H7R87_RS29610 (WP3S18E03_P40880) | 72362..72730 | + | 369 | WP_087829211 | type IV conjugative transfer system pilin TraA | - |
| H7R87_RS29615 (WP3S18E03_P40890) | 72744..73049 | + | 306 | WP_087829212 | type IV conjugative transfer system protein TraL | traL |
| H7R87_RS29620 (WP3S18E03_P40900) | 73069..73635 | + | 567 | WP_064373744 | type IV conjugative transfer system protein TraE | traE |
| H7R87_RS29625 (WP3S18E03_P40910) | 73622..74362 | + | 741 | WP_020804840 | type-F conjugative transfer system secretin TraK | traK |
| H7R87_RS29630 (WP3S18E03_P40920) | 74362..75786 | + | 1425 | WP_182949619 | F-type conjugal transfer pilus assembly protein TraB | traB |
| H7R87_RS29635 (WP3S18E03_P40930) | 75900..76469 | + | 570 | WP_182949616 | type IV conjugative transfer system lipoprotein TraV | traV |
| H7R87_RS30360 | 76625..77023 | + | 399 | WP_223177007 | hypothetical protein | - |
| H7R87_RS29645 (WP3S18E03_P40940) | 77052..77342 | + | 291 | WP_032426790 | hypothetical protein | - |
| H7R87_RS29650 (WP3S18E03_P40950) | 77366..77584 | + | 219 | WP_023316402 | hypothetical protein | - |
| H7R87_RS29655 (WP3S18E03_P40960) | 77585..77896 | + | 312 | WP_023316403 | hypothetical protein | - |
| H7R87_RS29660 (WP3S18E03_P40970) | 77963..78367 | + | 405 | WP_023316404 | hypothetical protein | - |
| H7R87_RS29665 (WP3S18E03_P40980) | 78364..78654 | + | 291 | WP_020323516 | hypothetical protein | - |
| H7R87_RS29670 (WP3S18E03_P40990) | 78662..79060 | + | 399 | WP_072156904 | hypothetical protein | - |
| H7R87_RS29675 (WP3S18E03_P41000) | 79132..81771 | + | 2640 | WP_020323518 | type IV secretion system protein TraC | virb4 |
| H7R87_RS29680 (WP3S18E03_P41010) | 81771..82160 | + | 390 | WP_020314627 | type-F conjugative transfer system protein TrbI | - |
| H7R87_RS29685 (WP3S18E03_P41020) | 82160..82786 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| H7R87_RS29690 (WP3S18E03_P41030) | 82830..83789 | + | 960 | WP_029497356 | conjugal transfer pilus assembly protein TraU | traU |
| H7R87_RS29695 (WP3S18E03_P41040) | 83802..84440 | + | 639 | WP_064182677 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| H7R87_RS29700 (WP3S18E03_P41050) | 84499..86454 | + | 1956 | WP_182949614 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| H7R87_RS29705 (WP3S18E03_P41060) | 86487..86768 | + | 282 | WP_032419932 | hypothetical protein | - |
| H7R87_RS29710 (WP3S18E03_P41070) | 86758..86985 | + | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| H7R87_RS29715 (WP3S18E03_P41080) | 86996..87322 | + | 327 | WP_020323521 | hypothetical protein | - |
| H7R87_RS29720 (WP3S18E03_P41090) | 87343..88095 | + | 753 | WP_136071131 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| H7R87_RS29725 (WP3S18E03_P41100) | 88550..88789 | + | 240 | WP_023316407 | type-F conjugative transfer system pilin chaperone TraQ | - |
| H7R87_RS29730 (WP3S18E03_P41110) | 88761..89318 | + | 558 | WP_182972117 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| H7R87_RS29735 | 89406..90103 | + | 698 | WP_227666874 | IS1 family transposase | - |
| H7R87_RS29740 (WP3S18E03_P41130) | 90141..90584 | + | 444 | WP_020804675 | F-type conjugal transfer protein TrbF | - |
| H7R87_RS29745 (WP3S18E03_P41140) | 90562..91941 | + | 1380 | WP_162495503 | conjugal transfer pilus assembly protein TraH | traH |
| H7R87_RS29750 (WP3S18E03_P41150) | 91941..94790 | + | 2850 | WP_032744228 | conjugal transfer mating-pair stabilization protein TraG | traG |
| H7R87_RS29755 (WP3S18E03_P41160) | 94796..95323 | + | 528 | WP_020803849 | hypothetical protein | - |
| H7R87_RS30365 (WP3S18E03_P41170) | 95506..96174 | + | 669 | WP_020803846 | hypothetical protein | - |
| H7R87_RS29765 | 96198..96458 | + | 261 | WP_020803850 | hypothetical protein | - |
| H7R87_RS29770 (WP3S18E03_P41180) | 96530..98842 | + | 2313 | WP_020803851 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 18027 | GenBank | NZ_AP022018 |
| Plasmid name | pWP3-S18-ESBL-03_4 | Incompatibility group | - |
| Plasmid size | 111708 bp | Coordinate of oriT [Strand] | 70277..70325 [-] |
| Host baterium | Klebsiella quasipneumoniae strain WP3-S18-ESBL-03 |
Cargo genes
| Drug resistance gene | blaGES-4, catB8, aac(6')-31, qacE, sul1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |