Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117587 |
| Name | oriT_pWP5-W18-ESBL-06_4 |
| Organism | Klebsiella quasipneumoniae strain WP5-W18-ESBL-06 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP022116 (19910..19958 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pWP5-W18-ESBL-06_4
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file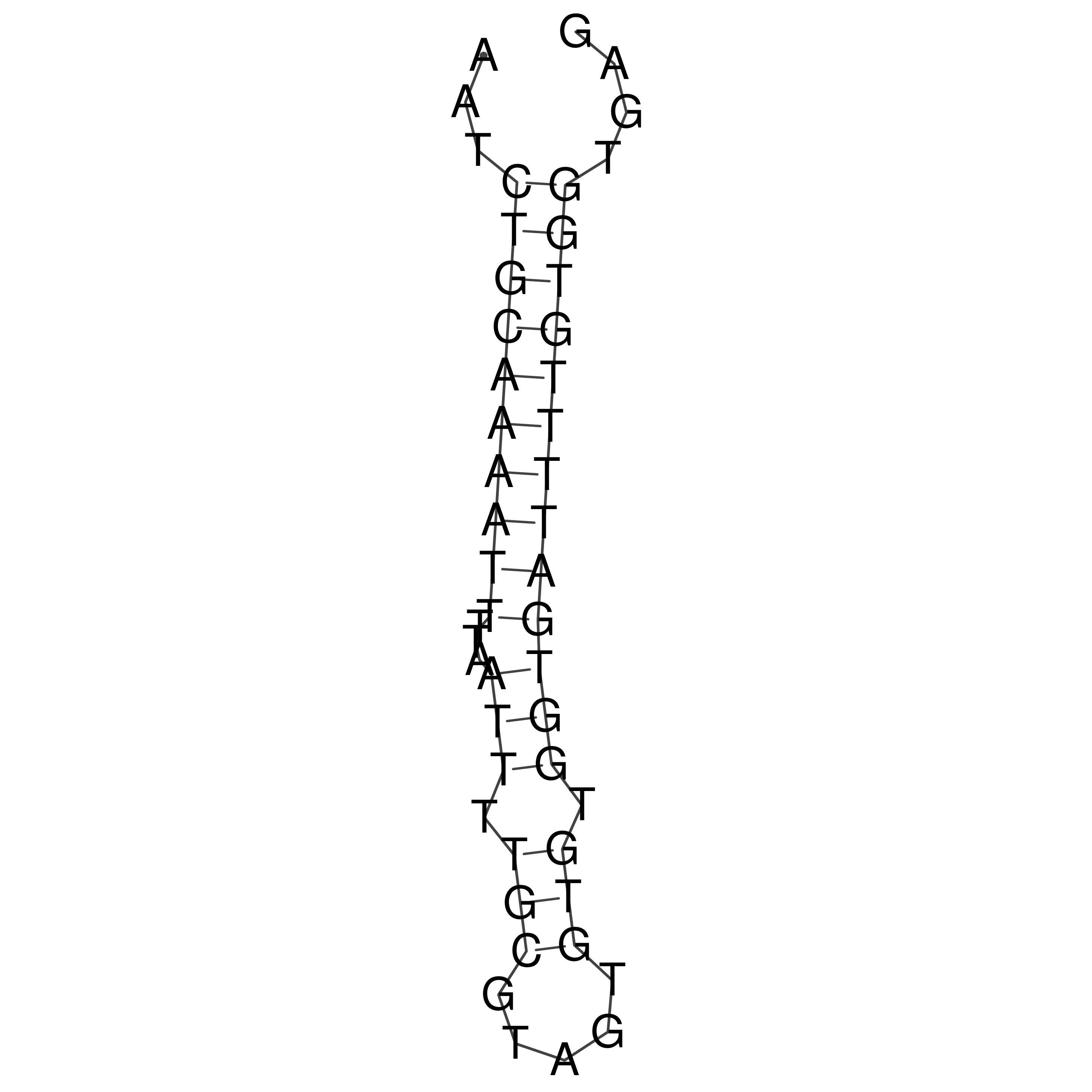
Relaxase
| ID | 11209 | GenBank | WP_182937559 |
| Name | traI_H7R83_RS29205_pWP5-W18-ESBL-06_4 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190358.14 Da Isoelectric Point: 6.3373
>WP_182937559.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGNHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMKDIEATIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHHALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPTQEKAIQK
ALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGNHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMKDIEATIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHHALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPTQEKAIQK
ALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6482 | GenBank | WP_004197733 |
| Name | WP_004197733_pWP5-W18-ESBL-06_4 |
UniProt ID | A0A0E3KB00 |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14835.90 Da Isoelectric Point: 4.6012
>WP_004197733.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Bacteria]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0E3KB00 |
T4CP
| ID | 13092 | GenBank | WP_009309880 |
| Name | traD_H7R83_RS29200_pWP5-W18-ESBL-06_4 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85571.58 Da Isoelectric Point: 5.1733
>WP_009309880.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22372..47275
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H7R83_RS29020 (WP5W18E06_P40200) | 17558..17905 | + | 348 | WP_022644944 | hypothetical protein | - |
| H7R83_RS29025 (WP5W18E06_P40210) | 17998..18144 | + | 147 | WP_022644943 | hypothetical protein | - |
| H7R83_RS29915 | 18192..18428 | + | 237 | Protein_22 | SAM-dependent DNA methyltransferase | - |
| H7R83_RS29035 (WP5W18E06_P40230) | 18777..19319 | + | 543 | WP_022644941 | antirestriction protein | - |
| H7R83_RS29040 (WP5W18E06_P40240) | 19343..19765 | - | 423 | WP_072205880 | transglycosylase SLT domain-containing protein | - |
| H7R83_RS29045 (WP5W18E06_P40250) | 20270..20659 | + | 390 | WP_004197733 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| H7R83_RS29050 (WP5W18E06_P40260) | 20898..21572 | + | 675 | WP_004197737 | hypothetical protein | - |
| H7R83_RS29055 | 21764..21928 | + | 165 | WP_032444506 | TraY domain-containing protein | - |
| H7R83_RS29060 (WP5W18E06_P40270) | 21990..22358 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| H7R83_RS29065 (WP5W18E06_P40280) | 22372..22677 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| H7R83_RS29070 (WP5W18E06_P40290) | 22697..23263 | + | 567 | WP_004194424 | type IV conjugative transfer system protein TraE | traE |
| H7R83_RS29075 (WP5W18E06_P40300) | 23250..23990 | + | 741 | WP_009309865 | type-F conjugative transfer system secretin TraK | traK |
| H7R83_RS29080 (WP5W18E06_P40310) | 23990..25414 | + | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| H7R83_RS29085 (WP5W18E06_P40320) | 25528..26112 | + | 585 | WP_009309868 | type IV conjugative transfer system lipoprotein TraV | traV |
| H7R83_RS29090 (WP5W18E06_P40330) | 26244..26654 | + | 411 | WP_009309869 | hypothetical protein | - |
| H7R83_RS29095 (WP5W18E06_P40340) | 26760..26978 | + | 219 | WP_004195468 | hypothetical protein | - |
| H7R83_RS29100 (WP5W18E06_P40350) | 26979..27290 | + | 312 | WP_004195465 | hypothetical protein | - |
| H7R83_RS29105 (WP5W18E06_P40360) | 27357..27761 | + | 405 | WP_004197817 | hypothetical protein | - |
| H7R83_RS29110 (WP5W18E06_P40370) | 28138..28536 | + | 399 | WP_023179972 | hypothetical protein | - |
| H7R83_RS29115 (WP5W18E06_P40380) | 28608..31247 | + | 2640 | WP_182937565 | type IV secretion system protein TraC | virb4 |
| H7R83_RS29120 (WP5W18E06_P40390) | 31247..31636 | + | 390 | WP_004197815 | type-F conjugative transfer system protein TrbI | - |
| H7R83_RS29125 (WP5W18E06_P40400) | 31636..32262 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| H7R83_RS29130 (WP5W18E06_P40410) | 32304..32693 | + | 390 | WP_004194992 | hypothetical protein | - |
| H7R83_RS29135 (WP5W18E06_P40420) | 32690..33679 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| H7R83_RS29140 (WP5W18E06_P40430) | 33692..34330 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| H7R83_RS29145 (WP5W18E06_P40440) | 34389..36344 | + | 1956 | WP_109043214 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| H7R83_RS29150 (WP5W18E06_P40450) | 36376..36630 | + | 255 | WP_004195500 | conjugal transfer protein TrbE | - |
| H7R83_RS29155 (WP5W18E06_P40460) | 36608..36856 | + | 249 | WP_004152675 | hypothetical protein | - |
| H7R83_RS29160 (WP5W18E06_P40470) | 36869..37195 | + | 327 | WP_004194451 | hypothetical protein | - |
| H7R83_RS29165 (WP5W18E06_P40480) | 37216..37968 | + | 753 | WP_182937567 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| H7R83_RS29170 (WP5W18E06_P40490) | 37979..38218 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| H7R83_RS29175 (WP5W18E06_P40500) | 38190..38762 | + | 573 | WP_009309876 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| H7R83_RS29180 (WP5W18E06_P40510) | 38755..39183 | + | 429 | WP_004152689 | hypothetical protein | - |
| H7R83_RS29185 (WP5W18E06_P40520) | 39170..40540 | + | 1371 | WP_009309877 | conjugal transfer pilus assembly protein TraH | traH |
| H7R83_RS29190 (WP5W18E06_P40530) | 40540..43413 | + | 2874 | WP_009309878 | conjugal transfer mating-pair stabilization protein TraG | traG |
| H7R83_RS29195 (WP5W18E06_P40550) | 44148..44840 | + | 693 | WP_182937557 | hypothetical protein | - |
| H7R83_RS29200 (WP5W18E06_P40560) | 44966..47275 | + | 2310 | WP_009309880 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 18020 | GenBank | NZ_AP022116 |
| Plasmid name | pWP5-W18-ESBL-06_4 | Incompatibility group | - |
| Plasmid size | 82271 bp | Coordinate of oriT [Strand] | 19910..19958 [-] |
| Host baterium | Klebsiella quasipneumoniae strain WP5-W18-ESBL-06 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |