Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117451 |
| Name | oriT_E2|unnamed1 |
| Organism | Lactiplantibacillus plantarum strain E2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP110243 (10274..10294 [-], 21 nt) |
| oriT length | 21 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 21 nt
>oriT_E2|unnamed1
AGTGGGGTGTAAGTGCGCATT
AGTGGGGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file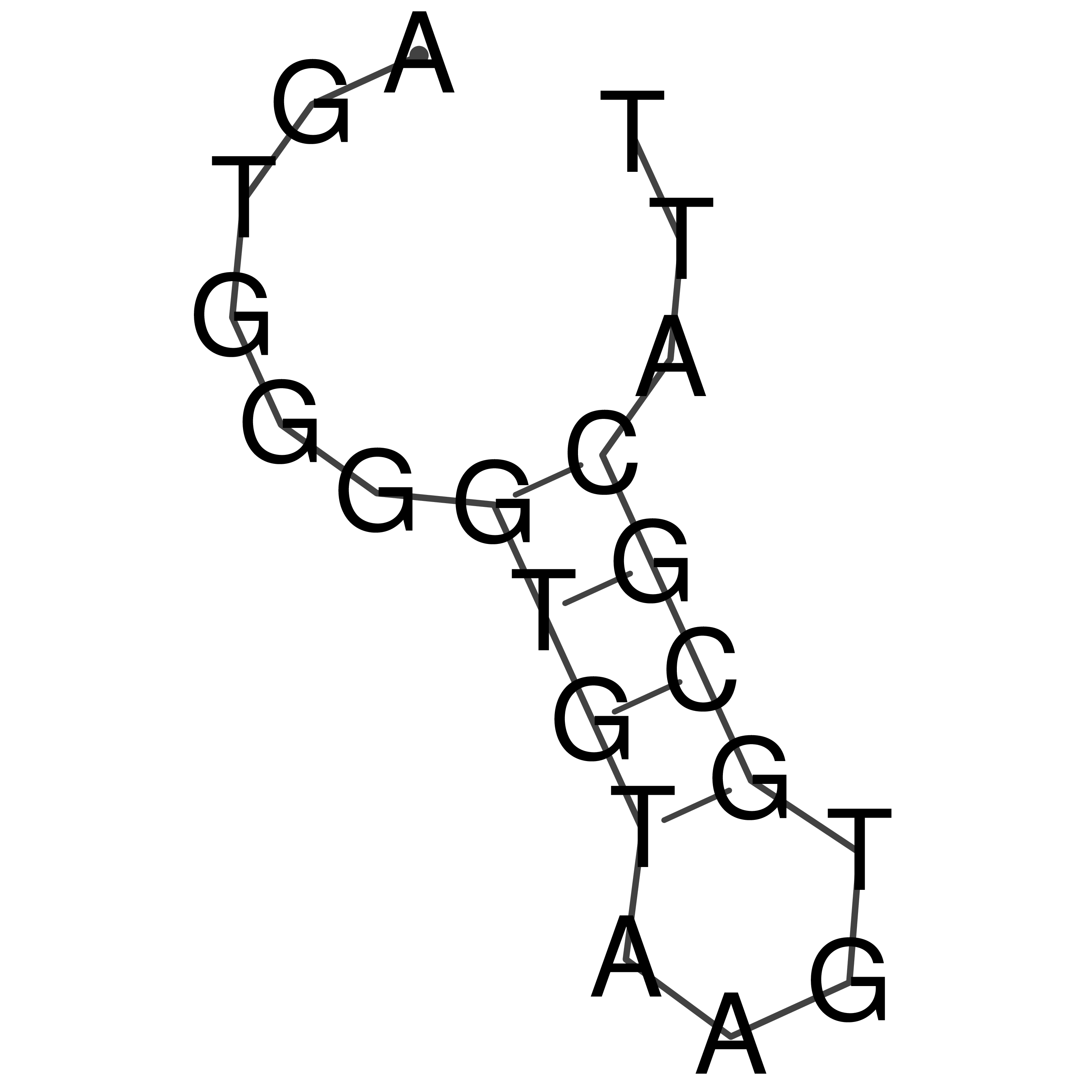
Relaxase
| ID | 11115 | GenBank | WP_264993783 |
| Name | mobQ_OLJ37_RS15635_E2|unnamed1 |
UniProt ID | _ |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80728.19 Da Isoelectric Point: 8.6531
>WP_264993783.1 MobQ family relaxase [Lactiplantibacillus plantarum]
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDKEGRHYFYAQSIMPESFILTPKNSPEWASDREQLWN
EVEKKDRKSNSRYAKEFNVALPVELSESEQKELLTKYVQENFVDEGMVADVAIHRDHADNPHAHVMLTNR
PFNPDGTWGIKSKKQYILDENGNKTYTGTSKYPKSRKILMVDWDKKEKIIEWRHNWAASVNQVLDQKNIP
DRISEKSFVDQGIDEVATQHEGINSKRYERKEFNQQVKNYRNTKASYKNNQEKAINRGHLDSLSKHFSFN
EKRLVNELGHELKTYISLESLDDKRRMLFNWKNSTLIKHAIGEDVTKKLLTINQQESSLKKADELLNKVV
DRTTKKLYPELDFEQTTQAERRELIKETESEQTIFKGSELNERLMNICDDLLTRQLLVFTKRPYVGWKLL
MQQEKEVKIELKYTLMIHDDSLESLEHVDQGLLEKYSSTEQQKITRAVKDLRTIMAVEQVIQTQYQEVLR
RTFPNGNFNELPMIKQEQAYTAVMYYDPALKPCKVETIAQWQENPPRVFTTQEHQQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKADPMIKNSQIEKIQKQLKEQQAKDDQYRKANMGHYQPLNYKPVSPDY
YLKTAFSDAIMTVLYARDEDYQRQKQERGLKETEWEMTKKQRQHQTRNRHEDGGRHL
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDKEGRHYFYAQSIMPESFILTPKNSPEWASDREQLWN
EVEKKDRKSNSRYAKEFNVALPVELSESEQKELLTKYVQENFVDEGMVADVAIHRDHADNPHAHVMLTNR
PFNPDGTWGIKSKKQYILDENGNKTYTGTSKYPKSRKILMVDWDKKEKIIEWRHNWAASVNQVLDQKNIP
DRISEKSFVDQGIDEVATQHEGINSKRYERKEFNQQVKNYRNTKASYKNNQEKAINRGHLDSLSKHFSFN
EKRLVNELGHELKTYISLESLDDKRRMLFNWKNSTLIKHAIGEDVTKKLLTINQQESSLKKADELLNKVV
DRTTKKLYPELDFEQTTQAERRELIKETESEQTIFKGSELNERLMNICDDLLTRQLLVFTKRPYVGWKLL
MQQEKEVKIELKYTLMIHDDSLESLEHVDQGLLEKYSSTEQQKITRAVKDLRTIMAVEQVIQTQYQEVLR
RTFPNGNFNELPMIKQEQAYTAVMYYDPALKPCKVETIAQWQENPPRVFTTQEHQQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKADPMIKNSQIEKIQKQLKEQQAKDDQYRKANMGHYQPLNYKPVSPDY
YLKTAFSDAIMTVLYARDEDYQRQKQERGLKETEWEMTKKQRQHQTRNRHEDGGRHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12986 | GenBank | WP_264993792 |
| Name | t4cp2_OLJ37_RS15695_E2|unnamed1 |
UniProt ID | _ |
| Length | 516 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 516 a.a. Molecular weight: 57999.14 Da Isoelectric Point: 4.8844
>WP_264993792.1 VirD4-like conjugal transfer protein, CD1115 family [Lactiplantibacillus plantarum]
MNGTILGIVDKKIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGVK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMK
HRSPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMSDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDQEVADFTSFSDFDLKEIGNAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNSANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPENQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNEYQGQPEASQETNFHQKVAKWEAKLKETAQKEAASEMSQKEEEQQQDNL
DSDLERVSNKDSQTEEPEEDDKNLIG
MNGTILGIVDKKIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGVK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMK
HRSPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMSDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDQEVADFTSFSDFDLKEIGNAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNSANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPENQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNEYQGQPEASQETNFHQKVAKWEAKLKETAQKEAASEMSQKEEEQQQDNL
DSDLERVSNKDSQTEEPEEDDKNLIG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13862..23951
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OLJ37_RS15615 (OLJ37_15615) | 8997..9187 | + | 191 | Protein_7 | hypothetical protein | - |
| OLJ37_RS15620 (OLJ37_15620) | 9202..9627 | + | 426 | WP_264993782 | zeta toxin family protein | - |
| OLJ37_RS15625 (OLJ37_15625) | 9617..9895 | - | 279 | WP_003645706 | hypothetical protein | - |
| OLJ37_RS15630 (OLJ37_15630) | 9912..10124 | - | 213 | WP_003645707 | hypothetical protein | - |
| OLJ37_RS15635 (OLJ37_15635) | 10396..12459 | + | 2064 | WP_264993783 | MobQ family relaxase | - |
| OLJ37_RS15640 (OLJ37_15640) | 12543..12854 | + | 312 | WP_264993784 | hypothetical protein | - |
| OLJ37_RS15645 (OLJ37_15645) | 12890..13504 | + | 615 | WP_264993785 | hypothetical protein | - |
| OLJ37_RS15650 (OLJ37_15650) | 13506..13841 | + | 336 | WP_024855764 | CagC family type IV secretion system protein | - |
| OLJ37_RS15655 (OLJ37_15655) | 13862..14224 | + | 363 | WP_195480458 | conjugal transfer protein | trsC |
| OLJ37_RS15660 (OLJ37_15660) | 14193..14852 | + | 660 | WP_068806586 | TrsD/TraD family conjugative transfer protein | trsD |
| OLJ37_RS15665 (OLJ37_15665) | 14864..16882 | + | 2019 | WP_264993786 | VirB4 family type IV secretion system protein | virb4 |
| OLJ37_RS15670 (OLJ37_15670) | 16875..18293 | + | 1419 | WP_264993787 | conjugal transfer protein | trsF |
| OLJ37_RS15675 (OLJ37_15675) | 18294..19451 | + | 1158 | WP_264993788 | phage tail tip lysozyme | prgK |
| OLJ37_RS15680 (OLJ37_15680) | 19466..20083 | + | 618 | WP_264993789 | hypothetical protein | - |
| OLJ37_RS15685 (OLJ37_15685) | 20070..20438 | + | 369 | WP_264993790 | thioredoxin family protein | - |
| OLJ37_RS15690 (OLJ37_15690) | 20439..20909 | + | 471 | WP_264993791 | conjugal transfer protein | trsJ |
| OLJ37_RS15695 (OLJ37_15695) | 21139..22689 | + | 1551 | WP_264993792 | VirD4-like conjugal transfer protein, CD1115 family | - |
| OLJ37_RS15700 (OLJ37_15700) | 22704..23093 | + | 390 | WP_264993793 | hypothetical protein | - |
| OLJ37_RS15705 (OLJ37_15705) | 23112..23951 | + | 840 | WP_060599186 | conjugal transfer protein TrbL family protein | trsL |
| OLJ37_RS15710 (OLJ37_15710) | 23966..24376 | + | 411 | WP_264993794 | hypothetical protein | - |
| OLJ37_RS15715 (OLJ37_15715) | 24383..26530 | + | 2148 | WP_264993795 | type IA DNA topoisomerase | - |
| OLJ37_RS15720 (OLJ37_15720) | 26687..28009 | - | 1323 | WP_264993796 | DNA (cytosine-5-)-methyltransferase | - |
| OLJ37_RS15725 (OLJ37_15725) | 28274..28687 | + | 414 | Protein_29 | IS30 family transposase | - |
Host bacterium
| ID | 17884 | GenBank | NZ_CP110243 |
| Plasmid name | E2|unnamed1 | Incompatibility group | - |
| Plasmid size | 54805 bp | Coordinate of oriT [Strand] | 10274..10294 [-] |
| Host baterium | Lactiplantibacillus plantarum strain E2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |