Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117151 |
| Name | oriT_pAtCFBP5507a |
| Organism | Agrobacterium salinitolerans strain CFBP5507 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP109970 (53059..53087 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pAtCFBP5507a
AGGGCGCAATATACGTCGCTATCGCGACG
AGGGCGCAATATACGTCGCTATCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file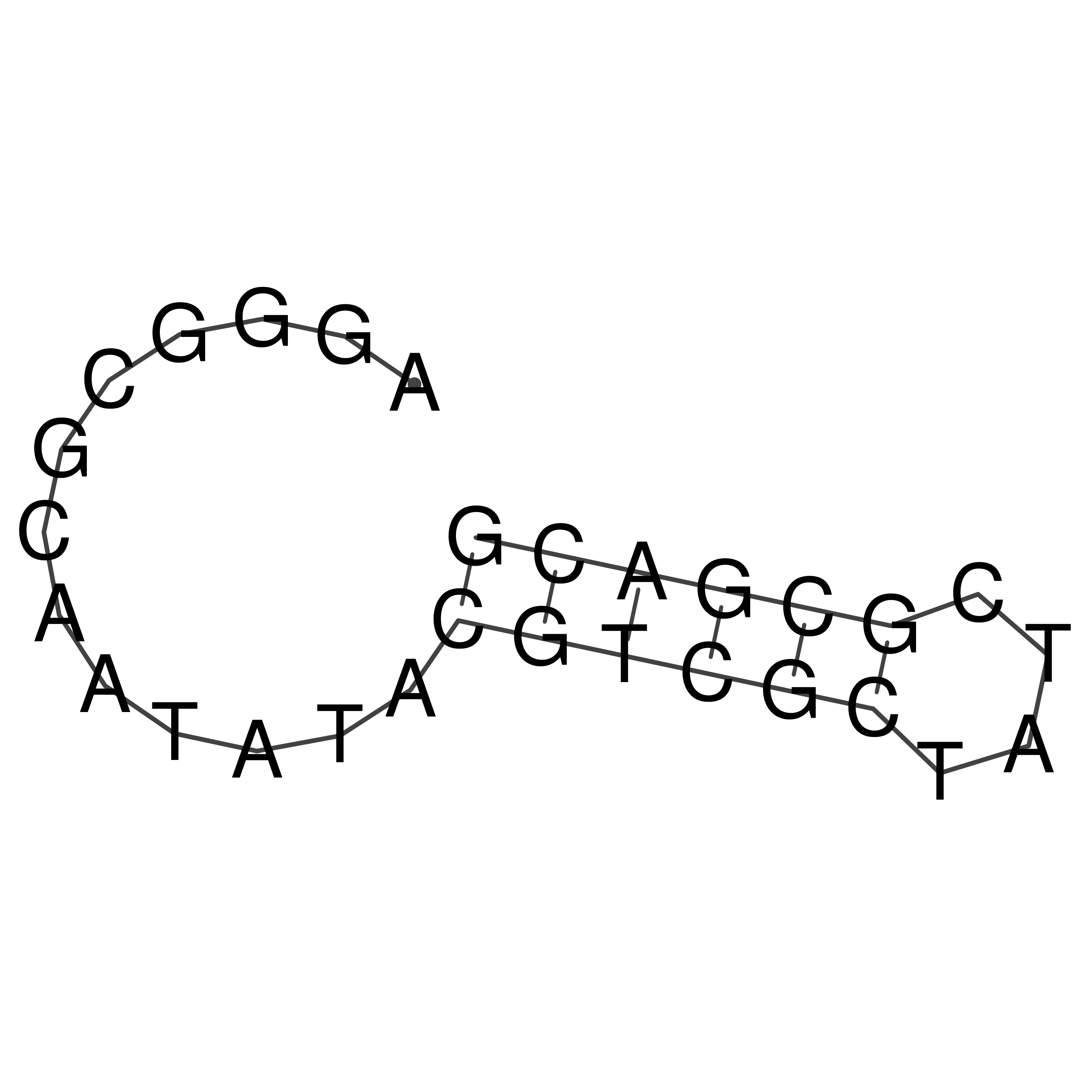
Relaxase
| ID | 10922 | GenBank | WP_137409261 |
| Name | traA_CFBP5507_RS24645_pAtCFBP5507a |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 170380.46 Da Isoelectric Point: 6.7532
>WP_137409261.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Agrobacterium]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPEGIPAWLMTAIDG
RTVAAASEALWNTVDAFETRADAQLARELIIALPEELTRAENIALVHEFVQDNFTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETMKAWKIVWADTANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQDMADRLLAEPELLLKQ
LSGERSTFDEKEIAKALHRYVDDPVDFANIRARLMASDDLVTLKPQVLDPETGKAAEPAVFTTRDMLRIE
YGMAQSARILSERKGFGVSSRKVAAAVDTVETADPEKRFRLDAEQVDAVRHVTGESGIAAVVGLAGAGKS
TLLAAARVAWESDGRRVVGAALAGKAAEGLEDSSGIKSRTLASWELAWASGRDLLERGNVLVIDEAGMVS
SQQMARALKAAEEAEAKVVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQREQWARDASRLFARGE
VEKGLDAYAQHGHLVEADTRDEIVARMVADWTQARNRAIEASVSEGREGRLRGDELLLLAHTNEDVKRLN
EALRSVMIDAGALGDCRSFRTERGEREFAAGDRIIFLENARFVEPRAKSFGPQYVKNGMLGTVVSTGDQR
GDALLSVRLDNGRDVVISENSYHNVDHGYAATIHKSQGSTVDRTLVLATGMMDQHLTYVSMTRHRDGADL
YAAKEDFEARPEWGRRPRADHAAGVTGELVETGNAKFRPDDEDADESPYADLKTDDGTVRRLWGVSLPKA
LEKAGVSESDTITLRKDGVEKVKVQVTVVDEQTGEKRTEEREVDRNVWTAEQKETAAARSKRIEMESHRP
ELFGRLVERLSRSGAKTTTLDFESEEGYGAQAQDFARRRGFDTLSKAAAGMEEGASRRLAWVAEKRELVA
KLWERASVALGFLIERERRASYNEDRAQTVAEGISISGHYLVPPTTQFARSVGEDARLAQLSSLAWMDRE
AILRPLLEKIYRDPVAALSTLNALASGGDVEPRTLAEDLAAAPDRLGRLRGSDLVVDGRIARNERADAMA
ALRELLPLARAHATEFRRQAERFGIREQQRRAHMSLSIPMLSKPATARLVEIERIRERGGDDAHKTAFAY
AVEDRQLVQEVKAVNDALTARFGWSAFTAKADVVAQRNIAERMPEDLAPECREKVLRLFAAVRRFSEEQH
LAEKQDRSKIVAGASIEIGKEAMPILPMLAAVTEFQTPVEEEAHNRAGAVAHYRHHRAALAETATLIWRD
PAGAVGKIEELVGKGFAGERIAAAVSNDPSAYGALRGSDRLMDWLLAPGRERKEALQAVPEAASRVRSLG
ASWATALDAETRAVTEERRRMVVAIPGLSQAAEDALRKLVADVNRQNKDKEKNTRRGVAAGSLEPRVVRE
FAEVSRALDERFGRNAILRGEKDTGNLVSPAQRRAFEAMRERLQVLQRAVRAESTQNIVAERQRRTIERG
RVFTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPEGIPAWLMTAIDG
RTVAAASEALWNTVDAFETRADAQLARELIIALPEELTRAENIALVHEFVQDNFTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETMKAWKIVWADTANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQDMADRLLAEPELLLKQ
LSGERSTFDEKEIAKALHRYVDDPVDFANIRARLMASDDLVTLKPQVLDPETGKAAEPAVFTTRDMLRIE
YGMAQSARILSERKGFGVSSRKVAAAVDTVETADPEKRFRLDAEQVDAVRHVTGESGIAAVVGLAGAGKS
TLLAAARVAWESDGRRVVGAALAGKAAEGLEDSSGIKSRTLASWELAWASGRDLLERGNVLVIDEAGMVS
SQQMARALKAAEEAEAKVVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQREQWARDASRLFARGE
VEKGLDAYAQHGHLVEADTRDEIVARMVADWTQARNRAIEASVSEGREGRLRGDELLLLAHTNEDVKRLN
EALRSVMIDAGALGDCRSFRTERGEREFAAGDRIIFLENARFVEPRAKSFGPQYVKNGMLGTVVSTGDQR
GDALLSVRLDNGRDVVISENSYHNVDHGYAATIHKSQGSTVDRTLVLATGMMDQHLTYVSMTRHRDGADL
YAAKEDFEARPEWGRRPRADHAAGVTGELVETGNAKFRPDDEDADESPYADLKTDDGTVRRLWGVSLPKA
LEKAGVSESDTITLRKDGVEKVKVQVTVVDEQTGEKRTEEREVDRNVWTAEQKETAAARSKRIEMESHRP
ELFGRLVERLSRSGAKTTTLDFESEEGYGAQAQDFARRRGFDTLSKAAAGMEEGASRRLAWVAEKRELVA
KLWERASVALGFLIERERRASYNEDRAQTVAEGISISGHYLVPPTTQFARSVGEDARLAQLSSLAWMDRE
AILRPLLEKIYRDPVAALSTLNALASGGDVEPRTLAEDLAAAPDRLGRLRGSDLVVDGRIARNERADAMA
ALRELLPLARAHATEFRRQAERFGIREQQRRAHMSLSIPMLSKPATARLVEIERIRERGGDDAHKTAFAY
AVEDRQLVQEVKAVNDALTARFGWSAFTAKADVVAQRNIAERMPEDLAPECREKVLRLFAAVRRFSEEQH
LAEKQDRSKIVAGASIEIGKEAMPILPMLAAVTEFQTPVEEEAHNRAGAVAHYRHHRAALAETATLIWRD
PAGAVGKIEELVGKGFAGERIAAAVSNDPSAYGALRGSDRLMDWLLAPGRERKEALQAVPEAASRVRSLG
ASWATALDAETRAVTEERRRMVVAIPGLSQAAEDALRKLVADVNRQNKDKEKNTRRGVAAGSLEPRVVRE
FAEVSRALDERFGRNAILRGEKDTGNLVSPAQRRAFEAMRERLQVLQRAVRAESTQNIVAERQRRTIERG
RVFTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12752 | GenBank | WP_137409258 |
| Name | traG_CFBP5507_RS24630_pAtCFBP5507a |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70624.24 Da Isoelectric Point: 9.2811
>WP_137409258.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Agrobacterium]
MTIRARPHLSLLLILMPITVTAFAAYVVGWRWPELAAGMSGETQYWFLRASPVPVLLFGPLAGLLTVWAL
PLHRRKPIAMATLICFLSVAGFYGLREFGRLSPSVESGAATWNQALSFIDMVAVVSAVVGFMATAASARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVIGERYRVDREIVHELPFDPNDPSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQNSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFRASDLTSGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIDADGAFERRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMTLLYQSVGQLERHFGKSGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKEMDTQAK
PNRFAKLRP
MTIRARPHLSLLLILMPITVTAFAAYVVGWRWPELAAGMSGETQYWFLRASPVPVLLFGPLAGLLTVWAL
PLHRRKPIAMATLICFLSVAGFYGLREFGRLSPSVESGAATWNQALSFIDMVAVVSAVVGFMATAASARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVIGERYRVDREIVHELPFDPNDPSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQNSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFRASDLTSGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIDADGAFERRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMTLLYQSVGQLERHFGKSGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKEMDTQAK
PNRFAKLRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6559..16140
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CFBP5507_RS24365 (CFBP5507_24365) | 2431..3702 | + | 1272 | WP_137409217 | plasmid replication protein RepC | - |
| CFBP5507_RS24370 (CFBP5507_24370) | 3901..4212 | - | 312 | WP_210239005 | hypothetical protein | - |
| CFBP5507_RS24375 (CFBP5507_24375) | 4575..4817 | + | 243 | WP_137409218 | hypothetical protein | - |
| CFBP5507_RS24380 (CFBP5507_24380) | 5561..5923 | - | 363 | WP_137409219 | transcriptional regulator | - |
| CFBP5507_RS24385 (CFBP5507_24385) | 6017..6556 | + | 540 | WP_137409220 | hypothetical protein | - |
| CFBP5507_RS24390 (CFBP5507_24390) | 6559..7239 | + | 681 | WP_137409221 | lytic transglycosylase domain-containing protein | virB1 |
| CFBP5507_RS24395 (CFBP5507_24395) | 7240..7539 | + | 300 | WP_137409222 | TrbC/VirB2 family protein | virB2 |
| CFBP5507_RS24400 (CFBP5507_24400) | 7542..7883 | + | 342 | WP_137409223 | type IV secretion system protein VirB3 | virB3 |
| CFBP5507_RS24405 (CFBP5507_24405) | 7876..10257 | + | 2382 | WP_137409224 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| CFBP5507_RS24410 (CFBP5507_24410) | 10257..10955 | + | 699 | WP_137409225 | P-type DNA transfer protein VirB5 | virB5 |
| CFBP5507_RS24415 (CFBP5507_24415) | 10952..11185 | + | 234 | WP_137409226 | EexN family lipoprotein | - |
| CFBP5507_RS24420 (CFBP5507_24420) | 11190..12125 | + | 936 | WP_137409227 | type IV secretion system protein | virB6 |
| CFBP5507_RS24425 (CFBP5507_24425) | 12159..12425 | + | 267 | WP_137409228 | hypothetical protein | - |
| CFBP5507_RS24430 (CFBP5507_24430) | 12429..13100 | + | 672 | WP_137409229 | virB8 family protein | virB8 |
| CFBP5507_RS24435 (CFBP5507_24435) | 13097..13948 | + | 852 | WP_170985381 | P-type conjugative transfer protein VirB9 | virB9 |
| CFBP5507_RS24440 (CFBP5507_24440) | 13961..15130 | + | 1170 | WP_137409230 | type IV secretion system protein VirB10 | virB10 |
| CFBP5507_RS24445 (CFBP5507_24445) | 15139..16140 | + | 1002 | WP_137409231 | P-type DNA transfer ATPase VirB11 | virB11 |
| CFBP5507_RS24450 (CFBP5507_24450) | 16417..17250 | - | 834 | WP_137409232 | SDR family oxidoreductase | - |
| CFBP5507_RS24455 (CFBP5507_24455) | 17323..17448 | - | 126 | WP_264673042 | hypothetical protein | - |
| CFBP5507_RS24460 (CFBP5507_24460) | 17543..18160 | + | 618 | WP_137409448 | TetR/AcrR family transcriptional regulator | - |
| CFBP5507_RS24465 (CFBP5507_24465) | 18624..18866 | - | 243 | WP_246666499 | hypothetical protein | - |
Host bacterium
| ID | 17584 | GenBank | NZ_CP109970 |
| Plasmid name | pAtCFBP5507a | Incompatibility group | - |
| Plasmid size | 343478 bp | Coordinate of oriT [Strand] | 53059..53087 [+] |
| Host baterium | Agrobacterium salinitolerans strain CFBP5507 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |