Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117139 |
| Name | oriT_pBE43_3 |
| Organism | Enterococcus faecalis strain BE43 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP110044 (18908..18945 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pBE43_3
ATACGAAGTAATGAAGTTACTGCGTATAAGTGCGCCCT
ATACGAAGTAATGAAGTTACTGCGTATAAGTGCGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file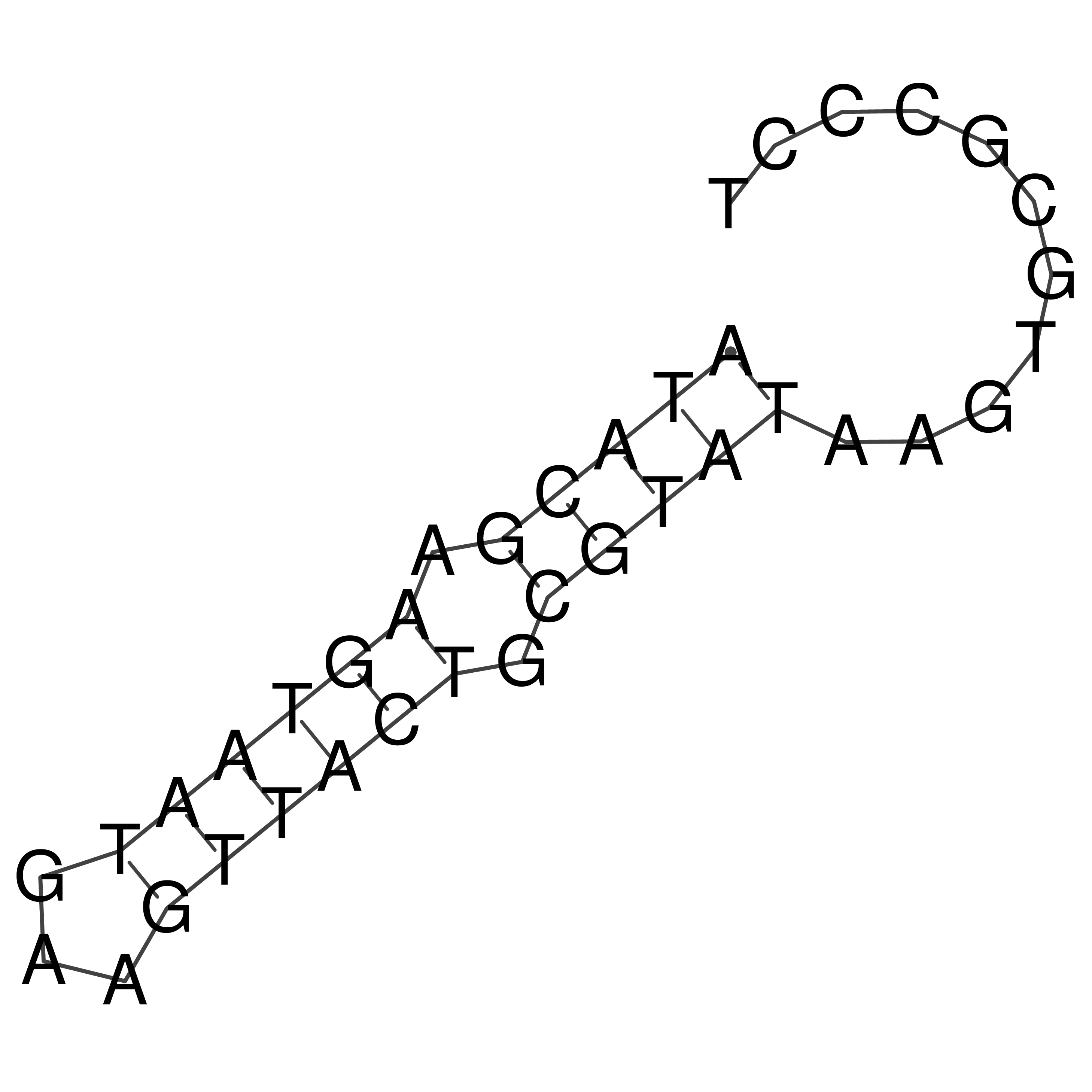
Relaxase
| ID | 10914 | GenBank | WP_319800784 |
| Name | mobQ_OLM01_RS14780_pBE43_3 |
UniProt ID | _ |
| Length | 661 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 661 a.a. Molecular weight: 77253.36 Da Isoelectric Point: 9.4365
>WP_319800784.1 MobQ family relaxase [Enterococcus faecalis]
MAIFHLSMTIAKRENGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNEWGRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLESQEKVLKQDQN
FTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKQNLKKVVNRNAYLPDSDKIELSKEIEKTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
MAIFHLSMTIAKRENGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNEWGRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLESQEKVLKQDQN
FTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKQNLKKVVNRNAYLPDSDKIELSKEIEKTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12745 | GenBank | WP_010723050 |
| Name | t4cp2_OLM01_RS14735_pBE43_3 |
UniProt ID | _ |
| Length | 551 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 551 a.a. Molecular weight: 63057.09 Da Isoelectric Point: 4.7399
>WP_010723050.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacilli]
MSSLLAESDGLILGKFPSGKVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELSMSDILSSFDEGIEENTANNDNSELPF
MSSLLAESDGLILGKFPSGKVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELSMSDILSSFDEGIEENTANNDNSELPF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4270..16450
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| OLM01_RS14675 (OLM01_14675) | 560..859 | - | 300 | WP_047342400 | hypothetical protein | - |
| OLM01_RS14680 (OLM01_14680) | 1225..1497 | - | 273 | WP_011266100 | helix-turn-helix transcriptional regulator | - |
| OLM01_RS14685 (OLM01_14685) | 1934..2236 | - | 303 | WP_141427078 | ATP-binding protein | - |
| OLM01_RS14690 (OLM01_14690) | 2389..2784 | - | 396 | WP_141427077 | Lrp/AsnC family transcriptional regulator | - |
| OLM01_RS14695 (OLM01_14695) | 2888..3325 | + | 438 | WP_069854469 | VOC family protein | - |
| OLM01_RS14700 (OLM01_14700) | 3497..3670 | - | 174 | WP_264526519 | hypothetical protein | - |
| OLM01_RS14705 (OLM01_14705) | 3897..3995 | + | 99 | WP_264526520 | type I toxin-antitoxin system Fst family toxin | - |
| OLM01_RS14710 (OLM01_14710) | 4270..4986 | - | 717 | WP_264526521 | LPXTG cell wall anchor domain-containing protein | prgC |
| OLM01_RS14715 (OLM01_14715) | 5046..5414 | - | 369 | WP_000516429 | hypothetical protein | - |
| OLM01_RS14720 (OLM01_14720) | 5443..6414 | - | 972 | WP_002425009 | hypothetical protein | - |
| OLM01_RS14725 (OLM01_14725) | 6431..7363 | - | 933 | WP_010750197 | conjugal transfer protein TrbL family protein | trsL |
| OLM01_RS14730 (OLM01_14730) | 7363..8286 | - | 924 | WP_264526522 | hypothetical protein | traP |
| OLM01_RS14735 (OLM01_14735) | 8304..9959 | - | 1656 | WP_010723050 | VirD4-like conjugal transfer protein, CD1115 family | - |
| OLM01_RS14740 (OLM01_14740) | 9952..10383 | - | 432 | WP_010723049 | hypothetical protein | - |
| OLM01_RS14745 (OLM01_14745) | 10388..10939 | - | 552 | WP_010723048 | hypothetical protein | - |
| OLM01_RS14750 (OLM01_14750) | 10952..12061 | - | 1110 | WP_264526523 | lysozyme family protein | orf14 |
| OLM01_RS14755 (OLM01_14755) | 12083..13435 | - | 1353 | WP_264526524 | conjugal transfer protein TraF | trsF |
| OLM01_RS14760 (OLM01_14760) | 13449..15410 | - | 1962 | WP_264526525 | VirB4 family type IV secretion system protein | virb4 |
| OLM01_RS14765 (OLM01_14765) | 15421..16050 | - | 630 | WP_319800785 | hypothetical protein | trsD |
| OLM01_RS14770 (OLM01_14770) | 16067..16450 | - | 384 | WP_002332651 | hypothetical protein | trsC |
| OLM01_RS14775 (OLM01_14775) | 16469..16801 | - | 333 | WP_000713503 | CagC family type IV secretion system protein | - |
| OLM01_RS14780 (OLM01_14780) | 16825..18810 | - | 1986 | WP_319800784 | MobQ family relaxase | - |
| OLM01_RS14785 (OLM01_14785) | 19102..19401 | + | 300 | WP_264526510 | hypothetical protein | - |
| OLM01_RS14790 (OLM01_14790) | 19404..19661 | + | 258 | WP_264526511 | hypothetical protein | - |
| OLM01_RS14795 (OLM01_14795) | 19684..19989 | - | 306 | WP_264526512 | hypothetical protein | - |
| OLM01_RS14800 (OLM01_14800) | 20023..20520 | - | 498 | WP_002300565 | DnaJ domain-containing protein | - |
| OLM01_RS14805 (OLM01_14805) | 20540..20857 | - | 318 | WP_002338433 | hypothetical protein | - |
Host bacterium
| ID | 17572 | GenBank | NZ_CP110044 |
| Plasmid name | pBE43_3 | Incompatibility group | - |
| Plasmid size | 28952 bp | Coordinate of oriT [Strand] | 18908..18945 [+] |
| Host baterium | Enterococcus faecalis strain BE43 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |