Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 117131 |
| Name | oriT_p36B2_p1 |
| Organism | Vagococcus fluvialis strain 36B2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP081462 (44290..44327 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 7..12, 16..21 (AGTAAC..GTTACT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_p36B2_p1
ATACGAAGTAACGAAGTTACTGCGTATAAGTGCGCCCT
ATACGAAGTAACGAAGTTACTGCGTATAAGTGCGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file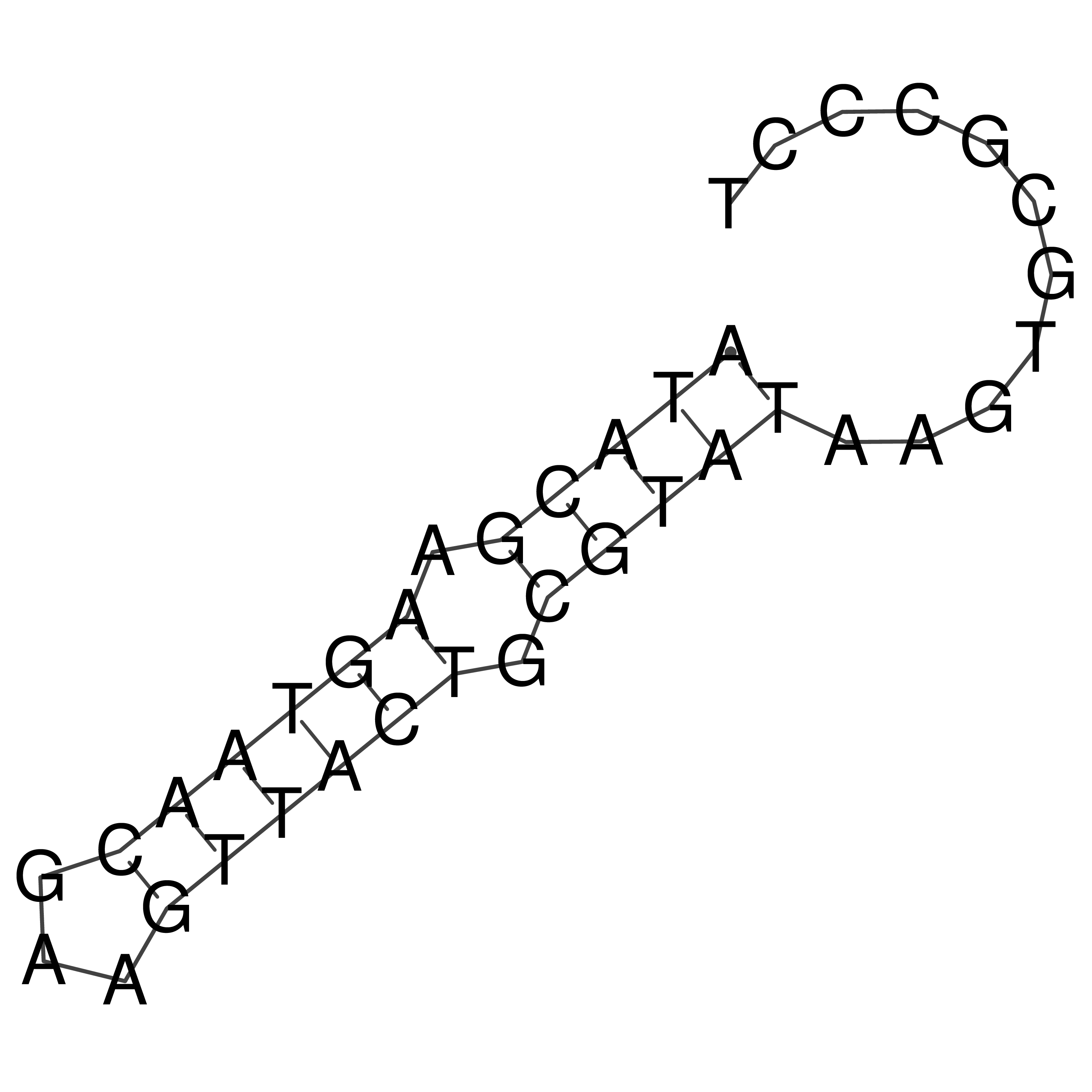
Relaxase
| ID | 10909 | GenBank | WP_319796307 |
| Name | mobQ_K5K99_RS14575_p36B2_p1 |
UniProt ID | _ |
| Length | 661 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 661 a.a. Molecular weight: 77196.18 Da Isoelectric Point: 9.1581
>WP_319796307.1 MobQ family relaxase [Vagococcus fluvialis]
MAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNDWGRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLENQEKVLKQDQN
FTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKQNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKENQSLQRNIPLR
MAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNDWGRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLENQEKVLKQDQN
FTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKQNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKENQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12740 | GenBank | WP_002425008 |
| Name | t4cp2_K5K99_RS14530_p36B2_p1 |
UniProt ID | _ |
| Length | 551 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 551 a.a. Molecular weight: 63127.22 Da Isoelectric Point: 4.7399
>WP_002425008.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacilli]
MTSLLAESDGLILGKFPSGKVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELLMSDILSSFDEGIEENTTNNDNSELPF
MTSLLAESDGLILGKFPSGKVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELLMSDILSSFDEGIEENTTNNDNSELPF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 29511..41832
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| K5K99_RS14480 (K5K99_14440) | 25659..26522 | - | 864 | WP_002339793 | helix-turn-helix domain-containing protein | - |
| K5K99_RS14485 (K5K99_14445) | 26705..27055 | + | 351 | WP_002339794 | hypothetical protein | - |
| K5K99_RS14490 (K5K99_14450) | 27261..27551 | + | 291 | WP_002339795 | hypothetical protein | - |
| K5K99_RS14495 (K5K99_14455) | 27589..28269 | + | 681 | WP_002331383 | IS6-like element IS1216 family transposase | - |
| K5K99_RS14500 (K5K99_14460) | 28434..29399 | - | 966 | Protein_29 | IS256 family transposase | - |
| K5K99_RS14505 (K5K99_14465) | 29511..30359 | - | 849 | WP_227259516 | LPXTG cell wall anchor domain-containing protein | prgC |
| K5K99_RS14510 (K5K99_14470) | 30419..30787 | - | 369 | WP_000516429 | hypothetical protein | - |
| K5K99_RS14515 (K5K99_14475) | 30816..31784 | - | 969 | WP_227259517 | hypothetical protein | - |
| K5K99_RS14520 (K5K99_14480) | 31801..32745 | - | 945 | WP_227259518 | conjugal transfer protein TrbL family protein | trsL |
| K5K99_RS14525 (K5K99_14485) | 32745..33668 | - | 924 | WP_227259519 | hypothetical protein | - |
| K5K99_RS14530 (K5K99_14490) | 33686..35341 | - | 1656 | WP_002425008 | VirD4-like conjugal transfer protein, CD1115 family | - |
| K5K99_RS14535 (K5K99_14495) | 35334..35765 | - | 432 | WP_002425007 | hypothetical protein | - |
| K5K99_RS14540 (K5K99_14500) | 35770..36321 | - | 552 | WP_002425006 | hypothetical protein | - |
| K5K99_RS14545 (K5K99_14505) | 36334..37443 | - | 1110 | WP_227259520 | lysozyme family protein | orf14 |
| K5K99_RS14550 (K5K99_14510) | 37465..38817 | - | 1353 | WP_227259521 | conjugal transfer protein TraF | trsF |
| K5K99_RS14555 (K5K99_14515) | 38831..40792 | - | 1962 | WP_002425003 | VirB4 family type IV secretion system protein | virb4 |
| K5K99_RS14560 (K5K99_14520) | 40803..41432 | - | 630 | WP_002325627 | hypothetical protein | trsD |
| K5K99_RS14565 (K5K99_14525) | 41449..41832 | - | 384 | WP_001083514 | glycosyl transferase | trsC |
| K5K99_RS14570 (K5K99_14530) | 41851..42183 | - | 333 | WP_000713503 | CagC family type IV secretion system protein | - |
| K5K99_RS14575 (K5K99_14535) | 42207..44192 | - | 1986 | WP_319796307 | MobQ family relaxase | - |
| K5K99_RS14580 (K5K99_14540) | 44484..44783 | + | 300 | WP_227259523 | hypothetical protein | - |
| K5K99_RS14585 (K5K99_14545) | 44786..45043 | + | 258 | WP_227259524 | hypothetical protein | - |
| K5K99_RS14590 (K5K99_14550) | 45066..45341 | - | 276 | WP_227259525 | hypothetical protein | - |
| K5K99_RS14595 (K5K99_14555) | 45326..45736 | - | 411 | WP_125395807 | hypothetical protein | - |
| K5K99_RS14600 (K5K99_14560) | 45875..46828 | + | 954 | Protein_49 | IS256 family transposase | - |
Host bacterium
| ID | 17564 | GenBank | NZ_CP081462 |
| Plasmid name | p36B2_p1 | Incompatibility group | - |
| Plasmid size | 48594 bp | Coordinate of oriT [Strand] | 44290..44327 [+] |
| Host baterium | Vagococcus fluvialis strain 36B2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |