Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116921 |
| Name | oriT_Teo12|p3_Teo12 |
| Organism | Shinella zoogloeoides strain Teo12 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP131133 (116843..116871 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_Teo12|p3_Teo12
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file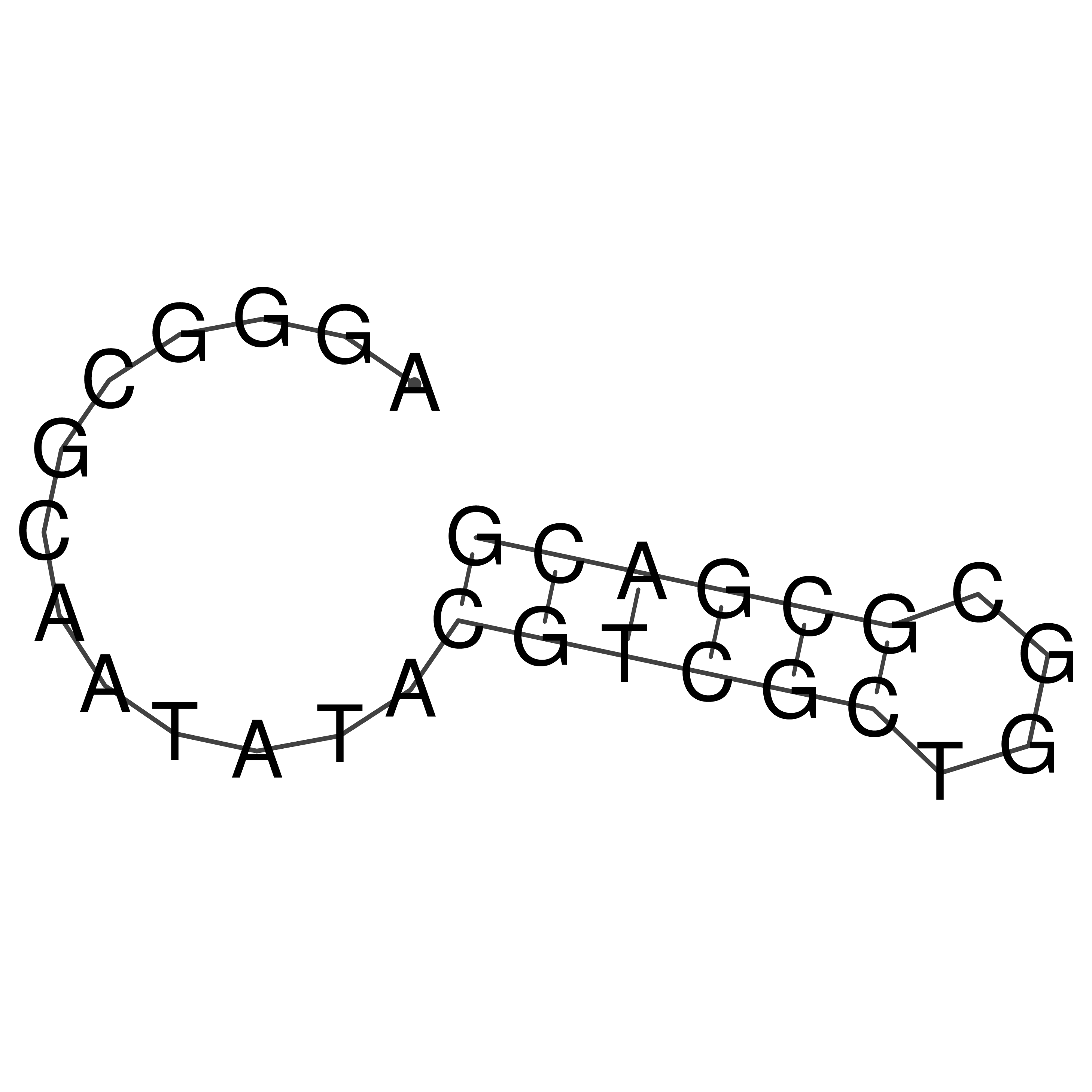
Relaxase
| ID | 10800 | GenBank | WP_318914513 |
| Name | traA_ShzoTeo12_RS28080_Teo12|p3_Teo12 |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170419.87 Da Isoelectric Point: 7.1789
>WP_318914513.1 Ti-type conjugative transfer relaxase TraA [Shinella zoogloeoides]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQVGTSFSYRGGATELKHEELALPDQVPAWLRNAIEG
KSVAGASEALWNAVEAFEARSDAQLARELIIALPEELTRAENIALVQEFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPKGKIVYKLWAGDKETMKAWKIGWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQDMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPEDFANIRARLMASDDLVLLKSQQVERQTGKVAEPAVFTTREILRIE
YDMAQSAEVLSKRTGFGIAESKVEAAVRQVETADPEKPFKLDREQVEAVRHVTGDSAIAAVVGLGGAGKS
TLLAAARVAWEGEHHRVFGAALAGKAAEGLEDSSGIKSRTLASWEIAWENGRDLLERDNVFVIDEAGMVS
SQQMARVLKTVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERIGSAELAGVRRQREGWARDASRLFARGK
VEEGLDAYAQRSRIVETEHRTEVVERIVADWSAARDEAIARSQTEGHGGRLRGDEMLVLAHTNDDVKRLN
SELRGVMNVAGALTGAREFQTERGVREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGTVVATGDLR
GGPLLSVRLDSGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDQAML
YAAREDFEPKPEWGRKPRADHAAGVTGELVETGEAKFRPDDDDVEESPYADVRTEDGSIHRVWGVSLPKA
LAKGGVSEGDTVTLRKDGVETVKVDVTITDLETGEKRTEKREVHRNVWTAELKETAFDRQERIERESHRP
ELFRQMVERLGRSGAKTTTLDFESEAGYQAHARDFARRRGIDLAAEVAAGMEEGAGRRLAWLAEKREQVA
KLWERASVALGFAIEREKSVSYNEERSETVAPRIPEDRPYLIPPTTRFARSIEEEARRAQLASERWKERE
AILRPVLEKIYRDPESALARLNVLASDAGMEPRRLAEDLVMRPQRLGRLQGSEHLVDGRAARDNRNAAMA
ALSDLLPLARSHATEFRRQAERFGIREEQRRGHMSLSIPALSKPAMARLVEIEAVRARGGPDAYKTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTEKADAVAERNMVERMPDDLASDRREKLARLFMVVRRFAEEQH
LAERKDRSKVVAGASVEPGKETSAGLPMLAAVTEFKTSVEDEARVRAPEVAHYGHNRAALVDTAGRIWRD
PAGAVDKIESLIVKGISGERIAAAVANDPAAYGALRGSDRIMDRLLAVGRERKDALQAVPEAESRVRSLG
ASYATAFANATQAITEERQRMGVAIPGLSPAADEALRALAAAMKKKDAKLNVAAGSLDPRIWEEFAGVSR
SLDLRFGRDAILRGEKDVVNRVSPGQRRPFEAMQERLKILQETVRMGSSQRIVAERQRRAIDRGRGVIR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQVGTSFSYRGGATELKHEELALPDQVPAWLRNAIEG
KSVAGASEALWNAVEAFEARSDAQLARELIIALPEELTRAENIALVQEFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPKGKIVYKLWAGDKETMKAWKIGWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQDMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPEDFANIRARLMASDDLVLLKSQQVERQTGKVAEPAVFTTREILRIE
YDMAQSAEVLSKRTGFGIAESKVEAAVRQVETADPEKPFKLDREQVEAVRHVTGDSAIAAVVGLGGAGKS
TLLAAARVAWEGEHHRVFGAALAGKAAEGLEDSSGIKSRTLASWEIAWENGRDLLERDNVFVIDEAGMVS
SQQMARVLKTVEDAGAKAVLVGDAMQLQPIQAGAAFRAISERIGSAELAGVRRQREGWARDASRLFARGK
VEEGLDAYAQRSRIVETEHRTEVVERIVADWSAARDEAIARSQTEGHGGRLRGDEMLVLAHTNDDVKRLN
SELRGVMNVAGALTGAREFQTERGVREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGTVVATGDLR
GGPLLSVRLDSGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDQAML
YAAREDFEPKPEWGRKPRADHAAGVTGELVETGEAKFRPDDDDVEESPYADVRTEDGSIHRVWGVSLPKA
LAKGGVSEGDTVTLRKDGVETVKVDVTITDLETGEKRTEKREVHRNVWTAELKETAFDRQERIERESHRP
ELFRQMVERLGRSGAKTTTLDFESEAGYQAHARDFARRRGIDLAAEVAAGMEEGAGRRLAWLAEKREQVA
KLWERASVALGFAIEREKSVSYNEERSETVAPRIPEDRPYLIPPTTRFARSIEEEARRAQLASERWKERE
AILRPVLEKIYRDPESALARLNVLASDAGMEPRRLAEDLVMRPQRLGRLQGSEHLVDGRAARDNRNAAMA
ALSDLLPLARSHATEFRRQAERFGIREEQRRGHMSLSIPALSKPAMARLVEIEAVRARGGPDAYKTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTEKADAVAERNMVERMPDDLASDRREKLARLFMVVRRFAEEQH
LAERKDRSKVVAGASVEPGKETSAGLPMLAAVTEFKTSVEDEARVRAPEVAHYGHNRAALVDTAGRIWRD
PAGAVDKIESLIVKGISGERIAAAVANDPAAYGALRGSDRIMDRLLAVGRERKDALQAVPEAESRVRSLG
ASYATAFANATQAITEERQRMGVAIPGLSPAADEALRALAAAMKKKDAKLNVAAGSLDPRIWEEFAGVSR
SLDLRFGRDAILRGEKDVVNRVSPGQRRPFEAMQERLKILQETVRMGSSQRIVAERQRRAIDRGRGVIR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12621 | GenBank | WP_318914510 |
| Name | traG_ShzoTeo12_RS28065_Teo12|p3_Teo12 |
UniProt ID | _ |
| Length | 640 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 640 a.a. Molecular weight: 70676.62 Da Isoelectric Point: 9.1336
>WP_318914510.1 Ti-type conjugative transfer system protein TraG [Shinella zoogloeoides]
MMVRAKPHPSLLLVLVPIVVSGFAFYVANWRWPELATGMSGKTQYWFLRTSPVPVLLFGPLAGLLAVWAL
PLHRRRPIAMAGFVCFLLMAGFYGLREVARLSPYVERGALSWDQALSFLDMVAVASAAVGFMAAAVAARI
SSVVPEPVKRAKHGTFGDADWLPMAAAARLFPDNGEIVVGERYRVDKELVHALPFDPNDPSTWGKGGTAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPIICLDPSTEVAPIVVEHRRKALRREVMVLDPT
NPIMGFNVLDGIETSTKKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEFADRRN
LRSLRQIVSEPETSVLAMLRNVQEHSASAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKSSEIAEGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIEADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKSSASRKSESLNFQRRPLIMPHEITQDMRKDEQIIIVQGHSPIRCGRAVYFRRKEMDAAAA
ANRFVKGGRP
MMVRAKPHPSLLLVLVPIVVSGFAFYVANWRWPELATGMSGKTQYWFLRTSPVPVLLFGPLAGLLAVWAL
PLHRRRPIAMAGFVCFLLMAGFYGLREVARLSPYVERGALSWDQALSFLDMVAVASAAVGFMAAAVAARI
SSVVPEPVKRAKHGTFGDADWLPMAAAARLFPDNGEIVVGERYRVDKELVHALPFDPNDPSTWGKGGTAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPIICLDPSTEVAPIVVEHRRKALRREVMVLDPT
NPIMGFNVLDGIETSTKKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEFADRRN
LRSLRQIVSEPETSVLAMLRNVQEHSASAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKSSEIAEGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIEADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKSSASRKSESLNFQRRPLIMPHEITQDMRKDEQIIIVQGHSPIRCGRAVYFRRKEMDAAAA
ANRFVKGGRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 19893..29494
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ShzoTeo12_RS27555 (ShzoTeo12_54440) | 15221..16588 | - | 1368 | WP_269704329 | tyrosine-type recombinase/integrase | - |
| ShzoTeo12_RS27560 (ShzoTeo12_54450) | 16585..17814 | - | 1230 | WP_269704327 | site-specific integrase | - |
| ShzoTeo12_RS27565 (ShzoTeo12_54460) | 17906..18226 | - | 321 | WP_318914534 | hypothetical protein | - |
| ShzoTeo12_RS27575 (ShzoTeo12_54490) | 19316..19492 | - | 177 | WP_318914536 | hypothetical protein | - |
| ShzoTeo12_RS27580 (ShzoTeo12_54500) | 19591..19749 | - | 159 | Protein_23 | transposase | - |
| ShzoTeo12_RS27585 (ShzoTeo12_54510) | 19893..20900 | - | 1008 | WP_318914537 | P-type DNA transfer ATPase VirB11 | virB11 |
| ShzoTeo12_RS27590 (ShzoTeo12_54520) | 20910..22082 | - | 1173 | WP_318914538 | type IV secretion system protein VirB10 | virB10 |
| ShzoTeo12_RS27595 (ShzoTeo12_54530) | 22096..22950 | - | 855 | WP_318914539 | P-type conjugative transfer protein VirB9 | virB9 |
| ShzoTeo12_RS27600 (ShzoTeo12_54540) | 22947..23618 | - | 672 | WP_318914540 | virB8 family protein | virB8 |
| ShzoTeo12_RS27605 (ShzoTeo12_54550) | 23623..23889 | - | 267 | WP_318914541 | hypothetical protein | - |
| ShzoTeo12_RS27610 (ShzoTeo12_54560) | 23923..24858 | - | 936 | WP_318914542 | type IV secretion system protein | virB6 |
| ShzoTeo12_RS27615 (ShzoTeo12_54570) | 24863..25096 | - | 234 | WP_318914543 | EexN family lipoprotein | - |
| ShzoTeo12_RS27620 (ShzoTeo12_54580) | 25093..25791 | - | 699 | WP_318914544 | P-type DNA transfer protein VirB5 | virB5 |
| ShzoTeo12_RS27625 (ShzoTeo12_54590) | 25791..28172 | - | 2382 | WP_318914545 | VirB4 family type IV secretion system protein | virb4 |
| ShzoTeo12_RS27630 (ShzoTeo12_54600) | 28165..28506 | - | 342 | WP_318914546 | type IV secretion system protein VirB3 | virB3 |
| ShzoTeo12_RS27635 (ShzoTeo12_54610) | 28512..28811 | - | 300 | WP_318914547 | TrbC/VirB2 family protein | virB2 |
| ShzoTeo12_RS27640 (ShzoTeo12_54620) | 28808..29494 | - | 687 | WP_318914548 | lytic transglycosylase domain-containing protein | virB1 |
| ShzoTeo12_RS27645 | 29497..30039 | - | 543 | WP_318914549 | hypothetical protein | - |
| ShzoTeo12_RS27650 | 30135..30491 | + | 357 | WP_318914550 | transcriptional regulator | - |
| ShzoTeo12_RS27655 (ShzoTeo12_54630) | 30848..32071 | + | 1224 | WP_318914551 | plasmid partitioning protein RepA | - |
| ShzoTeo12_RS27660 (ShzoTeo12_54640) | 32056..33123 | + | 1068 | WP_318914552 | plasmid partitioning protein RepB | - |
Host bacterium
| ID | 17354 | GenBank | NZ_CP131133 |
| Plasmid name | Teo12|p3_Teo12 | Incompatibility group | - |
| Plasmid size | 140013 bp | Coordinate of oriT [Strand] | 116843..116871 [+] |
| Host baterium | Shinella zoogloeoides strain Teo12 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | - |