Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116911 |
| Name | oriT_p-2 |
| Organism | Ensifer adhaerens strain E-60 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP084488 (1226356..1226396 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 20..25, 30..35 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_p-2
ATCAAAGTGCGCAATTATACGTCGCCGATGCGACGCTTTGC
ATCAAAGTGCGCAATTATACGTCGCCGATGCGACGCTTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file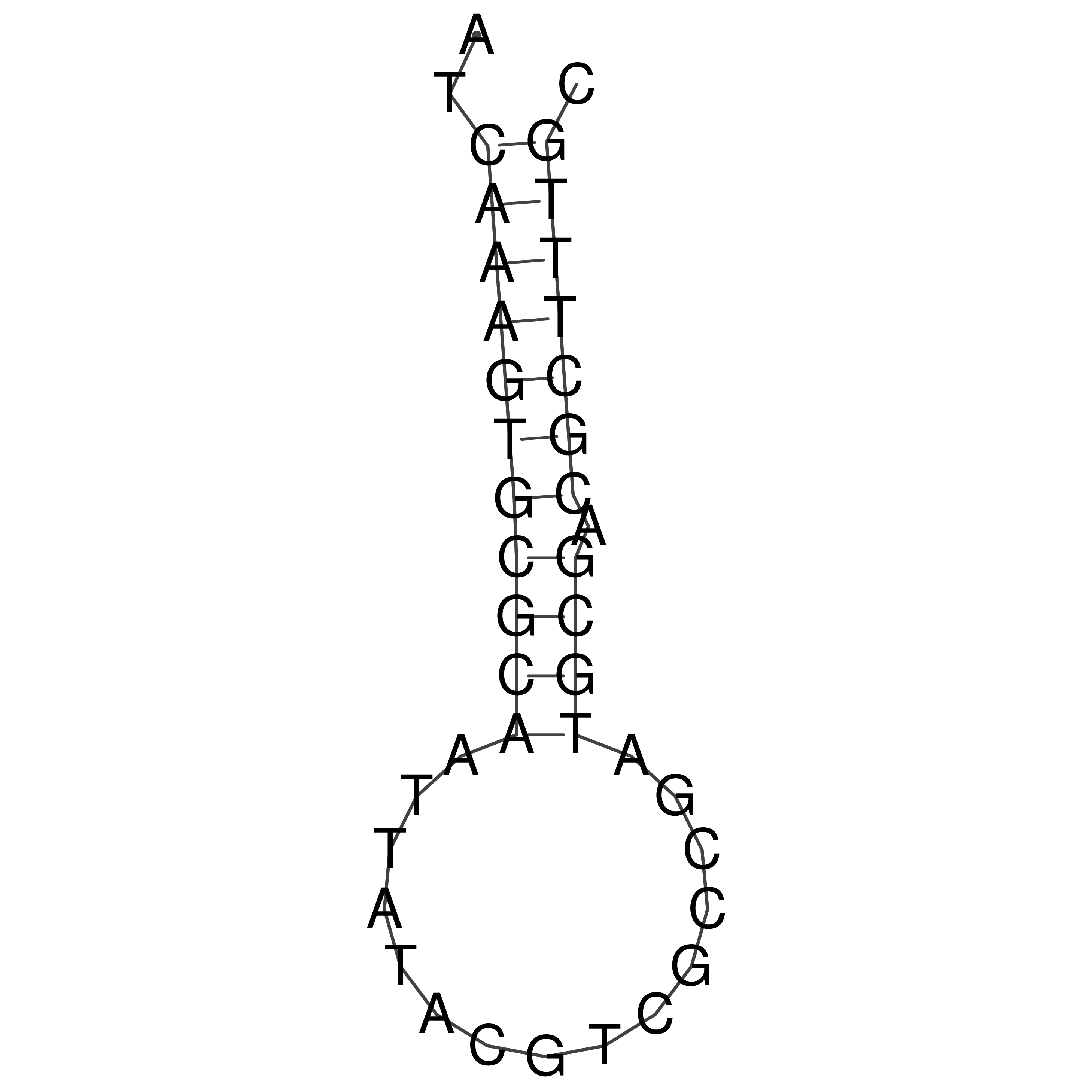
Relaxase
| ID | 10790 | GenBank | WP_218685710 |
| Name | traA_LDL63_RS32900_p-2 |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 122378.56 Da Isoelectric Point: 9.8756
>WP_218685710.1 Ti-type conjugative transfer relaxase TraA [Ensifer adhaerens]
MAVPHFSVSVVARGSGRSAVMSAAYRHCAKMDYEREARTIDYRAKQGLLHEEFAIPADAPAWLRSMIADR
PVSGAAEAFWNKVEQFEKRGDAQLAKDVTIALPLELTAEQNVALIRDFVEHYVTSRGMVADWVYHDAPGN
PHVHLMTTLRPLTENGFGGKKIAVLGVDGNPIRNDHGKIVYELWAGNLDDFNAFRDGWFACQNRHLGLAG
LGIRIDGRSFDKQGIELAPTIHLGVGTKAIERKSAADEQAASLERLALQEERRSENGRRIQGRPEIVLDL
ISREKSVFDERDIAKILHRYIDDPGLFQSLMARILQSPDALCLDRERLAFATGLRVPAKYATRELIRLEA
EMASRAVWLSRRSSHGVRDAVLKAAFTRHSRLSKEQISAIEHVAGPERIAAVIGRAGAGKTTMMKAAREA
WETAGYRVVGGALAGKAAEGLEKDAGIVSRTLSAWELRWKEGRNPLDNKTVFVLDEAGMVSSRQMALFVE
VATKAGAKLVLIGDPEQLQPIEAGAAFRAIADRIGYAELQTIYRQRERWMRDASLDLARGDVGKAVEAYR
ANGRIVASELKAQAVQNLIADWERDYDPTRTMLILAHLRRDVRMLNDMARARLVERGLLNDGFMFRTEDG
NRHFAPGDQIIFLKNEGSLGVKNGMLGKVVDAAQNRIVAEIGGGQHRRQVTVESRFYNNIDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLDVYYGQRSFAKAGGLTEMLSQKNSKETTLDYEKATFY
RQALHFAEARGLHLVNVARTIARDRLDWTVRQKQKLTDLAHRLASIGEQLGFAVGAKTMNAQRLKETRPM
VAGIATFPKSMEQSVIDKIEADPGLKKIWEDVSTRFHLVYAQPEAAFKAVNVGEMLKDDTVASATLAKVA
SQPDTYGALKGRTGILASRADKQDRERAECNVPALVQSLGDYVGQRSRIEQRYRTEEVALRRRVAIDIPE
LSAGARQTLERVRDAINRNELPAALGFALADRQVKAELDGFAKAVTERFGERTFLPLAAKDTNGETFKTI
AAGLKPGETAEVRLAWNMMRAAQQLSAHERTVEALKQAETLRQTKGQGLSLK
MAVPHFSVSVVARGSGRSAVMSAAYRHCAKMDYEREARTIDYRAKQGLLHEEFAIPADAPAWLRSMIADR
PVSGAAEAFWNKVEQFEKRGDAQLAKDVTIALPLELTAEQNVALIRDFVEHYVTSRGMVADWVYHDAPGN
PHVHLMTTLRPLTENGFGGKKIAVLGVDGNPIRNDHGKIVYELWAGNLDDFNAFRDGWFACQNRHLGLAG
LGIRIDGRSFDKQGIELAPTIHLGVGTKAIERKSAADEQAASLERLALQEERRSENGRRIQGRPEIVLDL
ISREKSVFDERDIAKILHRYIDDPGLFQSLMARILQSPDALCLDRERLAFATGLRVPAKYATRELIRLEA
EMASRAVWLSRRSSHGVRDAVLKAAFTRHSRLSKEQISAIEHVAGPERIAAVIGRAGAGKTTMMKAAREA
WETAGYRVVGGALAGKAAEGLEKDAGIVSRTLSAWELRWKEGRNPLDNKTVFVLDEAGMVSSRQMALFVE
VATKAGAKLVLIGDPEQLQPIEAGAAFRAIADRIGYAELQTIYRQRERWMRDASLDLARGDVGKAVEAYR
ANGRIVASELKAQAVQNLIADWERDYDPTRTMLILAHLRRDVRMLNDMARARLVERGLLNDGFMFRTEDG
NRHFAPGDQIIFLKNEGSLGVKNGMLGKVVDAAQNRIVAEIGGGQHRRQVTVESRFYNNIDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLDVYYGQRSFAKAGGLTEMLSQKNSKETTLDYEKATFY
RQALHFAEARGLHLVNVARTIARDRLDWTVRQKQKLTDLAHRLASIGEQLGFAVGAKTMNAQRLKETRPM
VAGIATFPKSMEQSVIDKIEADPGLKKIWEDVSTRFHLVYAQPEAAFKAVNVGEMLKDDTVASATLAKVA
SQPDTYGALKGRTGILASRADKQDRERAECNVPALVQSLGDYVGQRSRIEQRYRTEEVALRRRVAIDIPE
LSAGARQTLERVRDAINRNELPAALGFALADRQVKAELDGFAKAVTERFGERTFLPLAAKDTNGETFKTI
AAGLKPGETAEVRLAWNMMRAAQQLSAHERTVEALKQAETLRQTKGQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12610 | GenBank | WP_218685713 |
| Name | traG_LDL63_RS32885_p-2 |
UniProt ID | _ |
| Length | 641 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 641 a.a. Molecular weight: 69087.84 Da Isoelectric Point: 8.8831
>WP_218685713.1 Ti-type conjugative transfer system protein TraG [Ensifer adhaerens]
MTTDKLALATIPVALMTLTVSGMHGIEHWLSAFGKTDAARDTLGRIGVAAPYVSAAALGVIFLFASAGAV
SIKLAGWSVLFGAAATLLTATLREAARLSAFAGDVPPGMSVASYLDPATMVGAAAALLAGGYALRVALIG
NAAFRRPEPKRVKGQRALHGEADWMKMGQAAKLFSDTGGIVIGECYRVDQDSTAAEPFRADDRETWGAGG
KPPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSSEVAPMVCRHRKAAARFLRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIISDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDQ
ENQTLRQARTNLAEPEPKLRERLQSIYHNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFSTDALASGKTDVFINIDLKTLETHAGLARVIIGSFLNAIYNRDGQVKGRALFLLDEVARLGY
MQILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPDTADYISKRCGMTT
VEIDQVSRSFQSKGASRTRSKQLAARALIQPHEVLRMRTDEQIVFTAGNPPLRCGRAIWFRRKDMKACVD
ANRFHKIAQDV
MTTDKLALATIPVALMTLTVSGMHGIEHWLSAFGKTDAARDTLGRIGVAAPYVSAAALGVIFLFASAGAV
SIKLAGWSVLFGAAATLLTATLREAARLSAFAGDVPPGMSVASYLDPATMVGAAAALLAGGYALRVALIG
NAAFRRPEPKRVKGQRALHGEADWMKMGQAAKLFSDTGGIVIGECYRVDQDSTAAEPFRADDRETWGAGG
KPPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSSEVAPMVCRHRKAAARFLRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIISDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDQ
ENQTLRQARTNLAEPEPKLRERLQSIYHNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFSTDALASGKTDVFINIDLKTLETHAGLARVIIGSFLNAIYNRDGQVKGRALFLLDEVARLGY
MQILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPDTADYISKRCGMTT
VEIDQVSRSFQSKGASRTRSKQLAARALIQPHEVLRMRTDEQIVFTAGNPPLRCGRAIWFRRKDMKACVD
ANRFHKIAQDV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 12611 | GenBank | WP_218685701 |
| Name | t4cp2_LDL63_RS32970_p-2 |
UniProt ID | _ |
| Length | 817 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 817 a.a. Molecular weight: 91663.38 Da Isoelectric Point: 5.8697
>WP_218685701.1 conjugal transfer protein TrbE [Ensifer adhaerens]
MVALKQFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
NGWMIQVEAIRVPTVDYPLEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHVLILTHRPPESKKTALSK
YIYSDESSRKKTYADTVLAIFRNSVREIEQYFANTLSIQRMETREVSERGGERTARYDELLQFVRFCITG
ENHPIRLPDVPMYLDWIATAELQHGLTPTVENRFLGVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRALDQDAMSMVAETEDAIAQASSQLVAYGYYTPVVIL
FDDDRERLQEKAEAIRRLVQAEGFGARIETLNATDAFLGSLPGNWYCNVREPLINTSNLADLVPLNSVWS
GSPVAPCPFFPTNSPPLMQVATGSTPFRFNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEFAQIFA
FDKGNSLLPLTLATGGEHYEIGGDHEEGRALAFCPLFDLSTDGDRAWASEWIEMLVALQGTTITPDHRNA
ISRQLCLMAKATGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLTLGTFQTFEIEHLMNLGE
RHLVPVLTYLFRLIEKRLDGSPSLIVLDEAWLMLGQPVFRNKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVSGAIPKREYYVVTSEGRRLFDMALGPV
ALSFAGASGKEDLKRIRALKSEHGQNWPSHWLATRGVPDAASILNIA
MVALKQFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
NGWMIQVEAIRVPTVDYPLEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHVLILTHRPPESKKTALSK
YIYSDESSRKKTYADTVLAIFRNSVREIEQYFANTLSIQRMETREVSERGGERTARYDELLQFVRFCITG
ENHPIRLPDVPMYLDWIATAELQHGLTPTVENRFLGVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRALDQDAMSMVAETEDAIAQASSQLVAYGYYTPVVIL
FDDDRERLQEKAEAIRRLVQAEGFGARIETLNATDAFLGSLPGNWYCNVREPLINTSNLADLVPLNSVWS
GSPVAPCPFFPTNSPPLMQVATGSTPFRFNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEFAQIFA
FDKGNSLLPLTLATGGEHYEIGGDHEEGRALAFCPLFDLSTDGDRAWASEWIEMLVALQGTTITPDHRNA
ISRQLCLMAKATGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLTLGTFQTFEIEHLMNLGE
RHLVPVLTYLFRLIEKRLDGSPSLIVLDEAWLMLGQPVFRNKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVSGAIPKREYYVVTSEGRRLFDMALGPV
ALSFAGASGKEDLKRIRALKSEHGQNWPSHWLATRGVPDAASILNIA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1231508..1243523
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LDL63_RS32905 (LDL63_32905) | 1229847..1230341 | + | 495 | WP_256939196 | conjugative transfer signal peptidase TraF | - |
| LDL63_RS32910 (LDL63_32910) | 1230331..1231491 | + | 1161 | WP_218685708 | conjugal transfer protein TraB | - |
| LDL63_RS32915 (LDL63_32915) | 1231508..1232110 | + | 603 | WP_218685707 | TraH family protein | virB1 |
| LDL63_RS32920 (LDL63_32920) | 1232366..1232575 | - | 210 | WP_060529727 | helix-turn-helix domain-containing protein | - |
| LDL63_RS32925 (LDL63_32925) | 1232750..1233121 | + | 372 | WP_060585894 | transcriptional repressor TraM | - |
| LDL63_RS32930 (LDL63_32930) | 1233074..1233784 | - | 711 | WP_218685706 | autoinducer binding domain-containing protein | - |
| LDL63_RS32935 (LDL63_32935) | 1234049..1235332 | - | 1284 | WP_218685705 | IncP-type conjugal transfer protein TrbI | virB10 |
| LDL63_RS32940 (LDL63_32940) | 1235343..1235777 | - | 435 | WP_090428444 | conjugal transfer protein TrbH | - |
| LDL63_RS32945 (LDL63_32945) | 1235780..1236592 | - | 813 | WP_218685704 | P-type conjugative transfer protein TrbG | virB9 |
| LDL63_RS32950 (LDL63_32950) | 1236609..1237271 | - | 663 | WP_113293051 | conjugal transfer protein TrbF | virB8 |
| LDL63_RS32955 (LDL63_32955) | 1237287..1238468 | - | 1182 | WP_218685703 | P-type conjugative transfer protein TrbL | virB6 |
| LDL63_RS32960 (LDL63_32960) | 1238462..1238659 | - | 198 | WP_113293053 | entry exclusion protein TrbK | - |
| LDL63_RS32965 (LDL63_32965) | 1238656..1239459 | - | 804 | WP_218685702 | P-type conjugative transfer protein TrbJ | virB5 |
| LDL63_RS32970 (LDL63_32970) | 1239431..1241884 | - | 2454 | WP_218685701 | conjugal transfer protein TrbE | virb4 |
| LDL63_RS32975 (LDL63_32975) | 1241893..1242192 | - | 300 | WP_060529729 | conjugal transfer protein TrbD | virB3 |
| LDL63_RS32980 (LDL63_32980) | 1242185..1242556 | - | 372 | WP_060529730 | TrbC/VirB2 family protein | virB2 |
| LDL63_RS32985 (LDL63_32985) | 1242546..1243523 | - | 978 | WP_218685700 | P-type conjugative transfer ATPase TrbB | virB11 |
| LDL63_RS32990 (LDL63_32990) | 1243534..1244160 | - | 627 | WP_218685699 | acyl-homoserine-lactone synthase TraI | - |
| LDL63_RS32995 (LDL63_32995) | 1244518..1245750 | + | 1233 | WP_218685777 | plasmid partitioning protein RepA | - |
| LDL63_RS33000 (LDL63_33000) | 1245801..1246814 | + | 1014 | WP_218685698 | plasmid partitioning protein RepB | - |
Host bacterium
| ID | 17344 | GenBank | NZ_CP084488 |
| Plasmid name | p-2 | Incompatibility group | - |
| Plasmid size | 1568420 bp | Coordinate of oriT [Strand] | 1226356..1226396 [+] |
| Host baterium | Ensifer adhaerens strain E-60 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | silP, nia, arsC |
| Degradation gene | RPYSC3_16500, linJ, dxnH, LinG |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | AcrIIA7, AcrIIA1 |