Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116806 |
| Name | oriT_pCMPG5300.03 |
| Organism | Lactiplantibacillus plantarum CMPG5300 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CM002921 (24590..24627 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..7, 19..25 (ACACAAC..GTTGTGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pCMPG5300.03
ACACAACCAACTTTAGTTGTTGTGTGTAAGTGCGCATT
ACACAACCAACTTTAGTTGTTGTGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file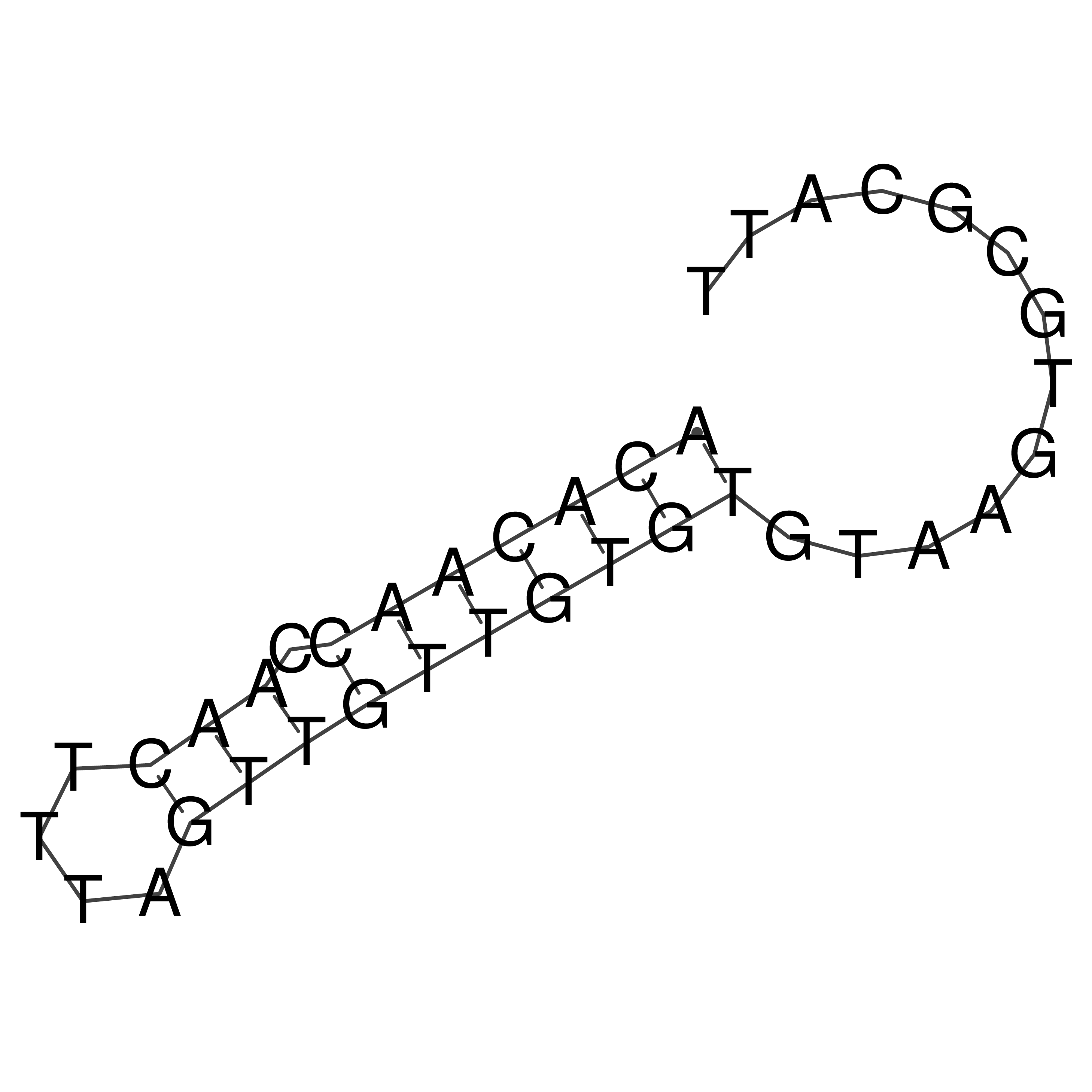
Relaxase
| ID | 10723 | GenBank | WP_047674898 |
| Name | mobQ_CMPG5300_RS15910_pCMPG5300.03 |
UniProt ID | _ |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80726.42 Da Isoelectric Point: 9.5573
>WP_047674898.1 MobQ family relaxase [Lactiplantibacillus plantarum]
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDKEGRHYFYARSVMPESFILTPKNAPEWASDRERLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNPDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKINQWRHNWAVSVNQVLAQKNIP
DRISEKSFIEQGIDDTPMQHEGINSKRHERKAFNQQVKAYRKSKAGYKNMQEKVINRGHLDSLSKHFSFN
EKRLVKELSHELKTYINLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLEKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTVFKGSELNERLMNIRDDLLTRQLLTFTKRPYVGWKLL
MQQEKKVKIELKYTLMIHDDSLESLEHVDQGLLEKYSPTEQQKITRAVKDLRTIMAVKQVIQTQYQEVLR
RAFPSGNFNELPMIKQEQAYTAVMYYDPALKPCKAETIAQWQETPPRVFNTQEHLQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKVDPTIKNSQIEKIQKQLKEQQAKDDQYRKVNMGHYQPLNYKPVSPSY
YLKTAFSNAIMTALYAHDEDYERQKQARGLKETEWEMTKKQRQHQTRNRHEDGGMHL
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDKEGRHYFYARSVMPESFILTPKNAPEWASDRERLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNPDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKINQWRHNWAVSVNQVLAQKNIP
DRISEKSFIEQGIDDTPMQHEGINSKRHERKAFNQQVKAYRKSKAGYKNMQEKVINRGHLDSLSKHFSFN
EKRLVKELSHELKTYINLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLEKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTVFKGSELNERLMNIRDDLLTRQLLTFTKRPYVGWKLL
MQQEKKVKIELKYTLMIHDDSLESLEHVDQGLLEKYSPTEQQKITRAVKDLRTIMAVKQVIQTQYQEVLR
RAFPSGNFNELPMIKQEQAYTAVMYYDPALKPCKAETIAQWQETPPRVFNTQEHLQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKVDPTIKNSQIEKIQKQLKEQQAKDDQYRKVNMGHYQPLNYKPVSPSY
YLKTAFSNAIMTALYAHDEDYERQKQARGLKETEWEMTKKQRQHQTRNRHEDGGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12523 | GenBank | WP_047674876 |
| Name | t4cp2_CMPG5300_RS15850_pCMPG5300.03 |
UniProt ID | _ |
| Length | 516 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 516 a.a. Molecular weight: 58037.36 Da Isoelectric Point: 5.0552
>WP_047674876.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Lactobacillaceae]
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRPPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMSDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVANFTSFSDFDLKEIGKAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKEKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEEQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETNFKNKVEKWKAKLKETAQKEVANKMSQKEEEQQQDNL
DSDLERVSNKDSQTEAPEEDDNNLLG
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRPPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMSDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVANFTSFSDFDLKEIGKAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKEKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEEQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETNFKNKVEKWKAKLKETAQKEVANKMSQKEEEQQQDNL
DSDLERVSNKDSQTEAPEEDDNNLLG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 10951..21039
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CMPG5300_RS17700 | 6713..6775 | + | 63 | WP_222843308 | putative holin-like toxin | - |
| CMPG5300_RS15820 (CMPG5300_3150) | 6928..8043 | - | 1116 | WP_047674872 | ArdC-like ssDNA-binding domain-containing protein | - |
| CMPG5300_RS15825 (CMPG5300_3151) | 8047..8262 | - | 216 | WP_007124624 | hypothetical protein | - |
| CMPG5300_RS15830 (CMPG5300_3152) | 8384..10519 | - | 2136 | WP_047674874 | DNA topoisomerase III | - |
| CMPG5300_RS15835 (CMPG5300_3153) | 10526..10936 | - | 411 | WP_012552821 | hypothetical protein | - |
| CMPG5300_RS15840 (CMPG5300_3154) | 10951..11790 | - | 840 | WP_003712161 | conjugal transfer protein TrbL family protein | trsL |
| CMPG5300_RS15845 (CMPG5300_3155) | 11809..12198 | - | 390 | WP_003712166 | hypothetical protein | - |
| CMPG5300_RS15850 (CMPG5300_3156) | 12213..13763 | - | 1551 | WP_047674876 | VirD4-like conjugal transfer protein, CD1115 family | - |
| CMPG5300_RS15855 (CMPG5300_3157) | 13993..14463 | - | 471 | WP_047674878 | hypothetical protein | trsJ |
| CMPG5300_RS15860 (CMPG5300_3158) | 14464..14832 | - | 369 | WP_047674881 | thioredoxin | - |
| CMPG5300_RS15865 (CMPG5300_3159) | 14819..15436 | - | 618 | WP_047674883 | hypothetical protein | - |
| CMPG5300_RS15870 (CMPG5300_3160) | 15450..16607 | - | 1158 | WP_047674886 | phage tail tip lysozyme | prgK |
| CMPG5300_RS15875 (CMPG5300_3161) | 16608..18026 | - | 1419 | WP_047674888 | conjugal transfer protein | trsF |
| CMPG5300_RS15880 (CMPG5300_3162) | 18019..20037 | - | 2019 | WP_047674890 | VirB4 family type IV secretion system protein | virb4 |
| CMPG5300_RS15885 (CMPG5300_3163) | 20049..20708 | - | 660 | WP_047674892 | TrsD/TraD family conjugative transfer protein | trsD |
| CMPG5300_RS15890 (CMPG5300_3164) | 20677..21039 | - | 363 | WP_003590770 | conjugal transfer protein | trsC |
| CMPG5300_RS15895 (CMPG5300_3165) | 21060..21395 | - | 336 | WP_047674894 | CagC family type IV secretion system protein | - |
| CMPG5300_RS15900 (CMPG5300_3166) | 21397..22011 | - | 615 | WP_047674896 | hypothetical protein | - |
| CMPG5300_RS15905 (CMPG5300_3167) | 22047..22358 | - | 312 | WP_021349367 | hypothetical protein | - |
| CMPG5300_RS15910 (CMPG5300_3168) | 22442..24505 | - | 2064 | WP_047674898 | MobQ family relaxase | - |
| CMPG5300_RS15915 (CMPG5300_3169) | 24759..24986 | + | 228 | WP_160945154 | hypothetical protein | - |
| CMPG5300_RS15920 (CMPG5300_3170) | 25009..25287 | + | 279 | WP_047674903 | hypothetical protein | - |
| CMPG5300_RS15925 (CMPG5300_3171) | 25277..25957 | - | 681 | WP_047674905 | zeta toxin family protein | - |
Host bacterium
| ID | 17239 | GenBank | NZ_CM002921 |
| Plasmid name | pCMPG5300.03 | Incompatibility group | - |
| Plasmid size | 27839 bp | Coordinate of oriT [Strand] | 24590..24627 [+] |
| Host baterium | Lactiplantibacillus plantarum CMPG5300 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |