Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116758 |
| Name | oriT_pl12113-1 |
| Organism | Levilactobacillus brevis strain TMW 1.2113 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP019751 (34606..34626 [+], 21 nt) |
| oriT length | 21 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 21 nt
>oriT_pl12113-1
AGTGGGGTGTAAGTGCGCATT
AGTGGGGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file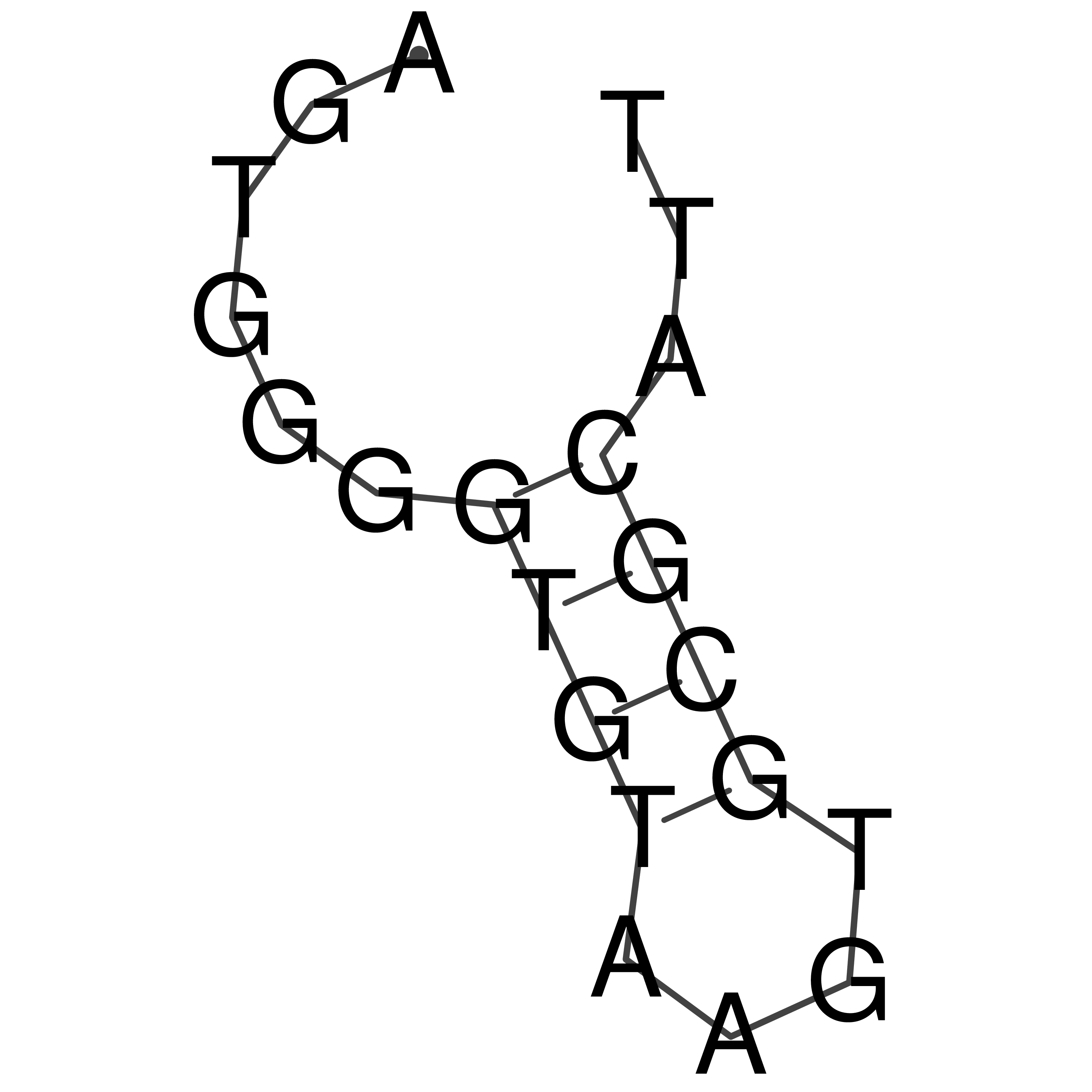
Relaxase
| ID | 10689 | GenBank | WP_085763780 |
| Name | mobQ_AZI10_RS12645_pl12113-1 |
UniProt ID | _ |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80339.13 Da Isoelectric Point: 9.4696
>WP_085763780.1 MobQ family relaxase [Levilactobacillus brevis]
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDQEGRHYFYARSVMPESFILTPKNAPAWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDQGMVADVAIHCDHQDNPHAHVMLTNR
PFNPDGTWGIKSKKQYILDENGNKMYTGTSKYPKSRKILMVDWDKKEKIIEWRHNWAVSVNQVLEQKNIP
DRISEKSFVEQGIADTPMQHEGINSKRHERKAFNQQVKKYRKTKANYKNNQEKVINRGHLDSLSKHFSFN
EKKVVKELSHELKTYISLESLDDKRRMLFNWKNSALIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELDFEQTTAAERRELIKETNSEQTIFKGSELNEHLMNIRDDLSARQLLAFTKRPYVGWKLL
MQQEKEAKIDLKYTLMIHSDSLESLEHVDKGLLEKYSPAEQQTITRAVKDLRTIMAVKQVIQTQYQEVLR
KAFPNGNFNELPMIKQEQAYTAVMYYDPALKPCKVETIAQWQENPPRVFSTQEHQQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKADPMIKNSQIEKIQKQLKEQQAKDDQYRKSQLAHYQPLNYKPVSPEY
YLKTAFSDAIMAALYARDEDYQRQRQAQGLKEAEWEMTKKQRQHQTRNRHEDGGMHL
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDQEGRHYFYARSVMPESFILTPKNAPAWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDQGMVADVAIHCDHQDNPHAHVMLTNR
PFNPDGTWGIKSKKQYILDENGNKMYTGTSKYPKSRKILMVDWDKKEKIIEWRHNWAVSVNQVLEQKNIP
DRISEKSFVEQGIADTPMQHEGINSKRHERKAFNQQVKKYRKTKANYKNNQEKVINRGHLDSLSKHFSFN
EKKVVKELSHELKTYISLESLDDKRRMLFNWKNSALIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELDFEQTTAAERRELIKETNSEQTIFKGSELNEHLMNIRDDLSARQLLAFTKRPYVGWKLL
MQQEKEAKIDLKYTLMIHSDSLESLEHVDKGLLEKYSPAEQQTITRAVKDLRTIMAVKQVIQTQYQEVLR
KAFPNGNFNELPMIKQEQAYTAVMYYDPALKPCKVETIAQWQENPPRVFSTQEHQQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKADPMIKNSQIEKIQKQLKEQQAKDDQYRKSQLAHYQPLNYKPVSPEY
YLKTAFSDAIMAALYARDEDYQRQRQAQGLKEAEWEMTKKQRQHQTRNRHEDGGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12493 | GenBank | WP_082265574 |
| Name | t4cp2_AZI10_RS12585_pl12113-1 |
UniProt ID | _ |
| Length | 516 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 516 a.a. Molecular weight: 58063.09 Da Isoelectric Point: 4.6825
>WP_082265574.1 VirD4-like conjugal transfer protein, CD1115 family [Levilactobacillus brevis]
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRAPDQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVAEFTSFSDFDLKEIGNDKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADNHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEEHSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPETSQETNFKDKVEKWKAKLKETAQKEAANEMSQTEEEQQQDNL
DSDLERVSNKDSQTEEPEEDDNNLLG
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRAPDQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVAEFTSFSDFDLKEIGNDKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADNHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEEHSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPETSQETNFKDKVEKWKAKLKETAQKEAANEMSQTEEEQQQDNL
DSDLERVSNKDSQTEEPEEDDNNLLG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 11788..31035
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AZI10_RS12515 (AZI10_12510) | 8019..8948 | + | 930 | WP_085763764 | IS30 family transposase | - |
| AZI10_RS12520 (AZI10_12515) | 9014..9901 | - | 888 | WP_085763765 | polysaccharide deacetylase family protein | - |
| AZI10_RS12530 (AZI10_12525) | 10199..11419 | - | 1221 | WP_085763767 | glycosyltransferase | - |
| AZI10_RS12535 (AZI10_12530) | 11788..13881 | + | 2094 | WP_191981953 | collagen-binding domain-containing protein | prgC |
| AZI10_RS12540 (AZI10_12535) | 13963..15453 | + | 1491 | WP_085763769 | glycosyltransferase | - |
| AZI10_RS12545 (AZI10_12540) | 15537..16593 | - | 1057 | Protein_13 | IS3 family transposase | - |
| AZI10_RS13345 (AZI10_12545) | 16934..16996 | + | 63 | WP_222843308 | putative holin-like toxin | - |
| AZI10_RS12555 (AZI10_12550) | 17152..18276 | - | 1125 | WP_085763771 | ArdC-like ssDNA-binding domain-containing protein | - |
| AZI10_RS12560 (AZI10_12555) | 18280..18495 | - | 216 | WP_085763772 | hypothetical protein | - |
| AZI10_RS12565 (AZI10_12560) | 18617..20752 | - | 2136 | WP_085763773 | type IA DNA topoisomerase | - |
| AZI10_RS12570 (AZI10_12565) | 20759..21169 | - | 411 | WP_085763774 | hypothetical protein | - |
| AZI10_RS12575 (AZI10_12570) | 21182..22021 | - | 840 | WP_085763775 | conjugal transfer protein TrbL family protein | trsL |
| AZI10_RS12580 (AZI10_12575) | 22040..22429 | - | 390 | WP_082265575 | hypothetical protein | - |
| AZI10_RS12585 (AZI10_12580) | 22444..23994 | - | 1551 | WP_082265574 | VirD4-like conjugal transfer protein, CD1115 family | - |
| AZI10_RS12590 (AZI10_12585) | 23996..24466 | - | 471 | WP_085763776 | conjugal transfer protein | trsJ |
| AZI10_RS12595 (AZI10_12590) | 24467..24835 | - | 369 | WP_076672537 | thioredoxin family protein | - |
| AZI10_RS12600 (AZI10_12595) | 24822..25439 | - | 618 | WP_082265572 | hypothetical protein | - |
| AZI10_RS12605 (AZI10_12600) | 25453..26606 | - | 1154 | Protein_25 | phage tail tip lysozyme | - |
| AZI10_RS12610 (AZI10_12605) | 26607..28025 | - | 1419 | WP_085763777 | type VII secretion protein EssB/YukC | trsF |
| AZI10_RS12615 (AZI10_12610) | 28018..30033 | - | 2016 | WP_085763778 | VirB4 family type IV secretion system protein | virb4 |
| AZI10_RS12620 (AZI10_12615) | 30045..30704 | - | 660 | WP_085763779 | TrsD/TraD family conjugative transfer protein | trsD |
| AZI10_RS12625 (AZI10_12620) | 30673..31035 | - | 363 | WP_060417621 | hypothetical protein | trsC |
| AZI10_RS12630 (AZI10_12625) | 31056..31394 | - | 339 | WP_082265510 | CagC family type IV secretion system protein | - |
| AZI10_RS12635 (AZI10_12630) | 31396..32010 | - | 615 | WP_082265511 | hypothetical protein | - |
| AZI10_RS12640 (AZI10_12635) | 32046..32357 | - | 312 | WP_025085230 | hypothetical protein | - |
| AZI10_RS12645 (AZI10_12640) | 32441..34504 | - | 2064 | WP_085763780 | MobQ family relaxase | - |
| AZI10_RS12650 (AZI10_12645) | 34757..34984 | + | 228 | WP_085763781 | hypothetical protein | - |
| AZI10_RS12655 (AZI10_12650) | 35006..35284 | + | 279 | WP_085763782 | hypothetical protein | - |
| AZI10_RS12660 (AZI10_12655) | 35274..35645 | - | 372 | Protein_36 | hypothetical protein | - |
Host bacterium
| ID | 17191 | GenBank | NZ_CP019751 |
| Plasmid name | pl12113-1 | Incompatibility group | - |
| Plasmid size | 41093 bp | Coordinate of oriT [Strand] | 34606..34626 [+] |
| Host baterium | Levilactobacillus brevis strain TMW 1.2113 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |