Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116710 |
| Name | oriT_pENKO-1 |
| Organism | Enterobacter kobei strain JCM 8580 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP024591 (63314..63368 [-], 55 nt) |
| oriT length | 55 nt |
| IRs (inverted repeats) | 21..27, 31..37 (GCAAGAC..GTCTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 55 nt
>oriT_pENKO-1
TGAAATTAAAGAATTAATCTGCAAGACAACGTCTTGCGTGGGGTGTGATGTTTTT
TGAAATTAAAGAATTAATCTGCAAGACAACGTCTTGCGTGGGGTGTGATGTTTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file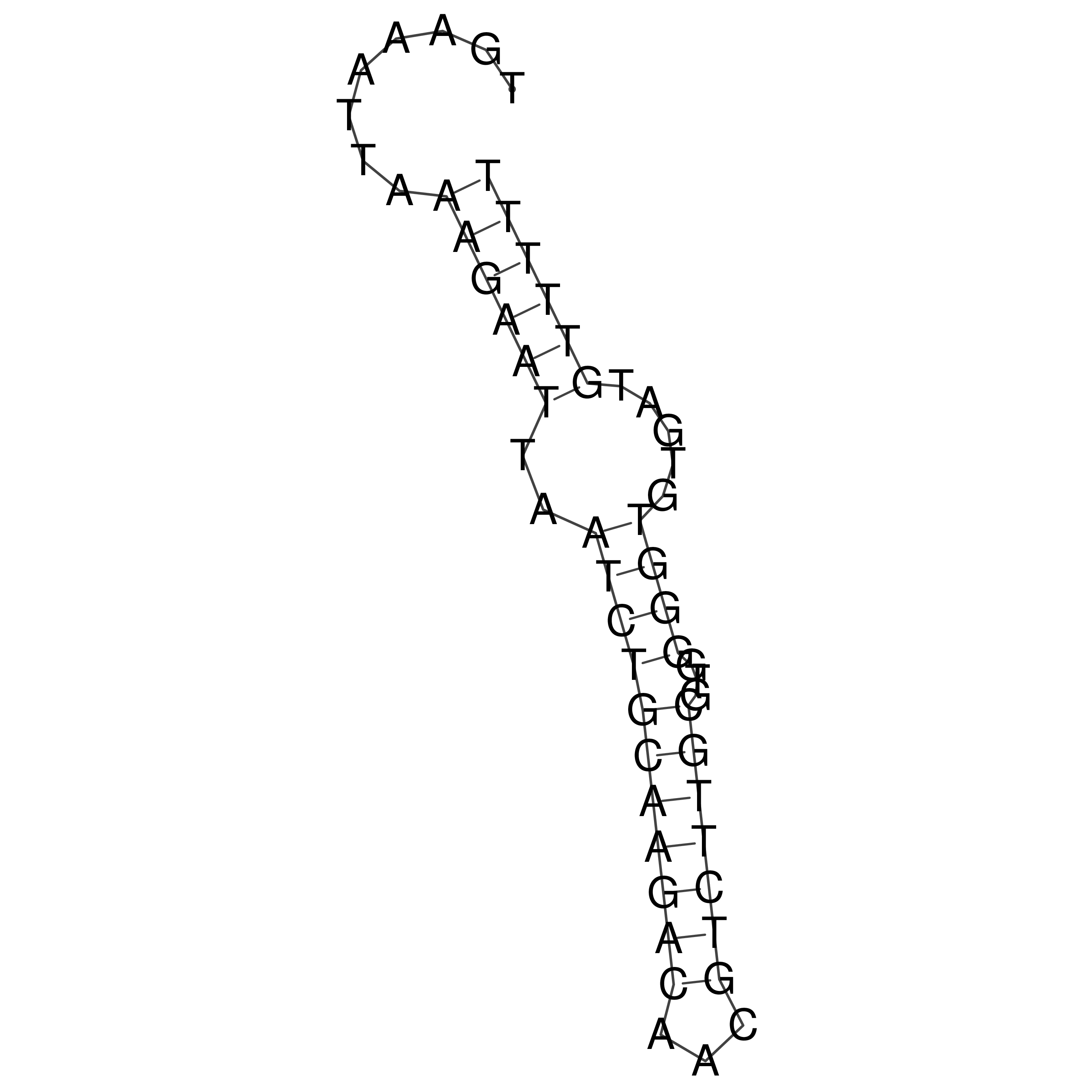
Relaxase
| ID | 10657 | GenBank | WP_088222281 |
| Name | traI_KI226_RS22525_pENKO-1 |
UniProt ID | _ |
| Length | 1830 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1830 a.a. Molecular weight: 198399.88 Da Isoelectric Point: 6.7335
>WP_088222281.1 conjugative transfer relaxase/helicase TraI [Enterobacter kobei]
MMSVAPVASAGTAAGYYSHSDNYYFLGNLQSHWLGEGARELGLDGPVRSDALTAVLEGRLPDGGRLGKEI
NGNHVHRPGHDLTFSAPKSVSILALIGGDKRMIDAHNHAVQVAAGYVEKLISARHTKDGVTSIVHTGKMV
AAAFTHDTSRNLDPLIHTHLLVANMTEYEGKWRALATDYIHNAGFIETVMKMQVTLGKIYRADLRQGVEA
LGHEVKEVGKHGMWEIKAIPDEVVEEYSSRGREVRGAVGADATLRSRDVAAKDTRRAKVDPSRIRLMERW
QTQMKEKGFDLKGYMDSVTPAESDTARPPAQIPSAADVQTRSQPDSTPEKVPARDRIPEAGKVTERDMAR
EEPSRPVTSTPPVMQAGTHAVSPVAQGAKAGPEKPHIRAEVSDAVRQAISHLSNDKTRFTWGELMLTTTE
FSDQLPDVAEVKMAIDASLKDGMIVPLDSEKGVFTSRIHLLDELSIQALSQERLKDGKVVSFARPEQYAP
RALEVVEKSPLVLMNAPTGVAGIRELTAQLTDISTAHGREVTVLASSAERASSLAKSDTLRDRLISRSHV
LSGEFSLKPQGTLIIEGAERLGLKETLVLLGEAREKDAQLVFLDSAGRQANGNAMSVLESAGVTRSRRTE
PAPGLETEVVSIADKRDRYAALASRFAELSAGSEPVTAVVLGQREQKHLTGLIRDALQNAGQLERDGVTV
EARTPVWVDNKTRRMPGTYRAGQVLEDRSDAKTTRHYVIDRIHEDTRVLSLIDSDGVLSRMKTSELTADW
RLYESENISISTGEQLIAVAGDKDAGLKAKDRLQVTGFSAGGIQVERDGLSLTLPADRPLYVKHAYVAAP
GGRDNDTGVVLAALNSRDISARTMNSLAQSGTRAEVFTAETQDRAETRLQRMNTSSSPVQLVRQLSGKDD
VSGAISALHDGVKSEAGLAVWRAINDQRGVTFSELTLLAKAAEYHPDIGAVGEHISTMVRQGDLLPVSVR
GEPALVARATWEMEKAIIRVIDAGKNTQEPLLEQVDPRLLDGLTAGQKAAARMVLGTTDQFIGIQGYAGV
GKTTQVKAVKAAIDSLPADGRPVLSGLAPTHQAVKELKDTGMPAQTVKSFLVEHDQQVNAGQKPDYRERV
FLIDESSMIGNQDTAAAYLAIQAGGGRAVSMGDIAQFEAVDAGAPFKLVQERSPMDVAIMKEIVRQKDVQ
LKGAVHDIIDNRIDAALRRIEAQPAGKIPRMAGATLPDSGIVTTEDAVGDIVRDWTERTPQARQNTLIIT
QLNADRQAVNAGIHAVLAERGELGEKSITVPVLEKISHTRHEFNKTAAWQAGMVVKRGDRYQDVVAVDKN
GSLVTVRDEDGRLGMVSPRELITGDVELFTRSTMTVNAGDELRFTATDRERGQTGNQRFTVQSVNDNGDI
VMKGAAGTKVINPAQVRAEQHIDYAWAVTGYGAQGASSEYVIALEGTEGGRKYLASQRAFYISASRAKEH
VQIYTDERSKWVAAMKQPEKEVKTAHDALQPETQRQQAKAIWAMGQPVTKTAIGRAWARHQSMAEHSLTA
RIIPATRRFPEPALALPLYDNNGKGAGLALISLVASPEGRMTQGDIRMVATEGASAAVLQRSQTGNTHVV
RHLSDALMAVREHPKDGVVWQTGEEKPSAHLMKVSRGVHQEEDAARIRAVSGLSTDITLPVTRDNPQREA
DQLSVLRAAEALRRAAEQERAGAVVDPEKLQHAVYKPESVTLPAEESIILPADLPQADTNPLFTPDSSTL
RKLAHDLTGGTEARIPAGMTRGENGTAPERAAAERASASKVVNDLAAAERDMVRQPVEGERGRAIEHEEY
AHTRTIQKER
MMSVAPVASAGTAAGYYSHSDNYYFLGNLQSHWLGEGARELGLDGPVRSDALTAVLEGRLPDGGRLGKEI
NGNHVHRPGHDLTFSAPKSVSILALIGGDKRMIDAHNHAVQVAAGYVEKLISARHTKDGVTSIVHTGKMV
AAAFTHDTSRNLDPLIHTHLLVANMTEYEGKWRALATDYIHNAGFIETVMKMQVTLGKIYRADLRQGVEA
LGHEVKEVGKHGMWEIKAIPDEVVEEYSSRGREVRGAVGADATLRSRDVAAKDTRRAKVDPSRIRLMERW
QTQMKEKGFDLKGYMDSVTPAESDTARPPAQIPSAADVQTRSQPDSTPEKVPARDRIPEAGKVTERDMAR
EEPSRPVTSTPPVMQAGTHAVSPVAQGAKAGPEKPHIRAEVSDAVRQAISHLSNDKTRFTWGELMLTTTE
FSDQLPDVAEVKMAIDASLKDGMIVPLDSEKGVFTSRIHLLDELSIQALSQERLKDGKVVSFARPEQYAP
RALEVVEKSPLVLMNAPTGVAGIRELTAQLTDISTAHGREVTVLASSAERASSLAKSDTLRDRLISRSHV
LSGEFSLKPQGTLIIEGAERLGLKETLVLLGEAREKDAQLVFLDSAGRQANGNAMSVLESAGVTRSRRTE
PAPGLETEVVSIADKRDRYAALASRFAELSAGSEPVTAVVLGQREQKHLTGLIRDALQNAGQLERDGVTV
EARTPVWVDNKTRRMPGTYRAGQVLEDRSDAKTTRHYVIDRIHEDTRVLSLIDSDGVLSRMKTSELTADW
RLYESENISISTGEQLIAVAGDKDAGLKAKDRLQVTGFSAGGIQVERDGLSLTLPADRPLYVKHAYVAAP
GGRDNDTGVVLAALNSRDISARTMNSLAQSGTRAEVFTAETQDRAETRLQRMNTSSSPVQLVRQLSGKDD
VSGAISALHDGVKSEAGLAVWRAINDQRGVTFSELTLLAKAAEYHPDIGAVGEHISTMVRQGDLLPVSVR
GEPALVARATWEMEKAIIRVIDAGKNTQEPLLEQVDPRLLDGLTAGQKAAARMVLGTTDQFIGIQGYAGV
GKTTQVKAVKAAIDSLPADGRPVLSGLAPTHQAVKELKDTGMPAQTVKSFLVEHDQQVNAGQKPDYRERV
FLIDESSMIGNQDTAAAYLAIQAGGGRAVSMGDIAQFEAVDAGAPFKLVQERSPMDVAIMKEIVRQKDVQ
LKGAVHDIIDNRIDAALRRIEAQPAGKIPRMAGATLPDSGIVTTEDAVGDIVRDWTERTPQARQNTLIIT
QLNADRQAVNAGIHAVLAERGELGEKSITVPVLEKISHTRHEFNKTAAWQAGMVVKRGDRYQDVVAVDKN
GSLVTVRDEDGRLGMVSPRELITGDVELFTRSTMTVNAGDELRFTATDRERGQTGNQRFTVQSVNDNGDI
VMKGAAGTKVINPAQVRAEQHIDYAWAVTGYGAQGASSEYVIALEGTEGGRKYLASQRAFYISASRAKEH
VQIYTDERSKWVAAMKQPEKEVKTAHDALQPETQRQQAKAIWAMGQPVTKTAIGRAWARHQSMAEHSLTA
RIIPATRRFPEPALALPLYDNNGKGAGLALISLVASPEGRMTQGDIRMVATEGASAAVLQRSQTGNTHVV
RHLSDALMAVREHPKDGVVWQTGEEKPSAHLMKVSRGVHQEEDAARIRAVSGLSTDITLPVTRDNPQREA
DQLSVLRAAEALRRAAEQERAGAVVDPEKLQHAVYKPESVTLPAEESIILPADLPQADTNPLFTPDSSTL
RKLAHDLTGGTEARIPAGMTRGENGTAPERAAAERASASKVVNDLAAAERDMVRQPVEGERGRAIEHEEY
AHTRTIQKER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12459 | GenBank | WP_088222282 |
| Name | traD_KI226_RS22520_pENKO-1 |
UniProt ID | _ |
| Length | 831 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 831 a.a. Molecular weight: 93179.46 Da Isoelectric Point: 5.0356
>WP_088222282.1 type IV conjugative transfer system coupling protein TraD [Enterobacter kobei]
MSFAGKNLTQGGQMTAYRWRMFIQVNNWIGFWVLILFVLIASSLFLWRTPQEMLDNGALYWFANLNQSFI
DLMPAGAGRFYDVHYYHAPTQTNYVLKMTLVQIYGDPYMNAMGTQVLEQLQWAALASGIFCTGVFAAISW
YISRIGKQESEDQYISGMQLTDKPADVNRLLRQNGELSDLRVGELHMVQRAEVMNYLIHGTIGVGKSTII
RWLLDYIRKRGDRAIIYDSGCTFTETHYDPRTDIILNAHDDRCANWQMWGECVDAVDYDNMAASLIPVEG
DSDPFWVSSSRTIYADLAIRMSADPDRSIEKFLKTLLSLSMQSLREYLANTPAANLVEEKIEKTAISIRS
VVTNYAKALRYLQGLDDGVKPPFTIREWMTTEKYDNSWLFISTQARHRKSVRPLISLWVSLATLMLQSMG
ENSDRRVWFIFDELPSLQRIPEFNETLAEGRKFGGCFVIGVQNMAQLVHAYGRELAKSMFDLLNTRMYGR
SPSAEMAEIVQKELGNQRKREIREQNSYGLDTVRDGISLGKDKVNNPIVDYEQIMRLPNLNFYVRLPGEY
PVVRLGLKYKKLVKRNAGLIERNIRDVLSPELEKVIQENERAATAAGLNFPTGDEVLENSNVATDTVPAV
APKPVQVKAESKPARTEVAKAEATASAKPLSVIPVSETAPEPETAPDQSNISDNVIVLKPLNVHKNPNLP
EREITASAVSRQPVTTHEAAGQDDLSSVTPALAVLRKFRQQAAGAASNEGDEDVSGGGESMSLDMHVSEL
ADGGLELSTAGTHEPGDDHPADHRASKSLAQDEENILLHRHPDDPGYSEYDLNYDDGERDL
MSFAGKNLTQGGQMTAYRWRMFIQVNNWIGFWVLILFVLIASSLFLWRTPQEMLDNGALYWFANLNQSFI
DLMPAGAGRFYDVHYYHAPTQTNYVLKMTLVQIYGDPYMNAMGTQVLEQLQWAALASGIFCTGVFAAISW
YISRIGKQESEDQYISGMQLTDKPADVNRLLRQNGELSDLRVGELHMVQRAEVMNYLIHGTIGVGKSTII
RWLLDYIRKRGDRAIIYDSGCTFTETHYDPRTDIILNAHDDRCANWQMWGECVDAVDYDNMAASLIPVEG
DSDPFWVSSSRTIYADLAIRMSADPDRSIEKFLKTLLSLSMQSLREYLANTPAANLVEEKIEKTAISIRS
VVTNYAKALRYLQGLDDGVKPPFTIREWMTTEKYDNSWLFISTQARHRKSVRPLISLWVSLATLMLQSMG
ENSDRRVWFIFDELPSLQRIPEFNETLAEGRKFGGCFVIGVQNMAQLVHAYGRELAKSMFDLLNTRMYGR
SPSAEMAEIVQKELGNQRKREIREQNSYGLDTVRDGISLGKDKVNNPIVDYEQIMRLPNLNFYVRLPGEY
PVVRLGLKYKKLVKRNAGLIERNIRDVLSPELEKVIQENERAATAAGLNFPTGDEVLENSNVATDTVPAV
APKPVQVKAESKPARTEVAKAEATASAKPLSVIPVSETAPEPETAPDQSNISDNVIVLKPLNVHKNPNLP
EREITASAVSRQPVTTHEAAGQDDLSSVTPALAVLRKFRQQAAGAASNEGDEDVSGGGESMSLDMHVSEL
ADGGLELSTAGTHEPGDDHPADHRASKSLAQDEENILLHRHPDDPGYSEYDLNYDDGERDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 65355..88427
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KI226_RS22370 (ENKO_43770) | 60478..60705 | + | 228 | WP_088222226 | hypothetical protein | - |
| KI226_RS22375 (ENKO_43790) | 61375..61677 | + | 303 | WP_140419642 | hypothetical protein | - |
| KI226_RS22380 (ENKO_43800) | 61737..62558 | + | 822 | WP_088222307 | DUF932 domain-containing protein | - |
| KI226_RS22385 | 62594..63118 | - | 525 | WP_088222313 | lytic transglycosylase domain-containing protein | - |
| KI226_RS22390 (ENKO_43820) | 63745..64137 | + | 393 | WP_088222306 | relaxosome protein TraM | - |
| KI226_RS22395 (ENKO_43830) | 64177..64809 | - | 633 | WP_088222305 | Arc family DNA-binding protein | - |
| KI226_RS22400 (ENKO_43840) | 65005..65352 | + | 348 | WP_088222304 | type IV conjugative transfer system pilin TraA | - |
| KI226_RS22405 (ENKO_43850) | 65355..65657 | + | 303 | WP_088222303 | type IV conjugative transfer system protein TraL | traL |
| KI226_RS22410 (ENKO_43860) | 65661..66242 | + | 582 | WP_088222302 | TraE/TraK family type IV conjugative transfer system protein | traE |
| KI226_RS22415 (ENKO_43870) | 66255..66995 | + | 741 | WP_088222301 | type-F conjugative transfer system secretin TraK | traK |
| KI226_RS22420 (ENKO_43880) | 66992..68383 | + | 1392 | WP_088222300 | F-type conjugal transfer pilus assembly protein TraB | traB |
| KI226_RS22425 (ENKO_43890) | 68425..68970 | + | 546 | WP_088222299 | type IV conjugative transfer system lipoprotein TraV | traV |
| KI226_RS22430 (ENKO_43900) | 68970..71579 | + | 2610 | WP_088222298 | type IV secretion system protein TraC | virb4 |
| KI226_RS22435 (ENKO_43910) | 71561..71986 | + | 426 | WP_088222297 | type-F conjugative transfer system protein TrbI | - |
| KI226_RS22440 (ENKO_43920) | 71983..72618 | + | 636 | WP_088222296 | type-F conjugative transfer system protein TraW | traW |
| KI226_RS22445 (ENKO_43930) | 72678..72959 | + | 282 | WP_254915068 | hypothetical protein | - |
| KI226_RS22450 (ENKO_43940) | 73111..73320 | + | 210 | WP_129364011 | hypothetical protein | - |
| KI226_RS22455 (ENKO_43950) | 73388..73729 | + | 342 | WP_088222294 | hypothetical protein | - |
| KI226_RS22460 (ENKO_43960) | 73714..74718 | + | 1005 | WP_088222293 | conjugal transfer pilus assembly protein TraU | traU |
| KI226_RS22465 (ENKO_43970) | 74726..75343 | + | 618 | WP_088222292 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| KI226_RS22470 (ENKO_43980) | 75340..77160 | + | 1821 | WP_088222291 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| KI226_RS22475 (ENKO_43990) | 77160..77942 | + | 783 | WP_088222290 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| KI226_RS22480 (ENKO_44000) | 78030..78254 | + | 225 | WP_140419641 | hypothetical protein | - |
| KI226_RS22485 (ENKO_44010) | 78244..78843 | + | 600 | WP_088222288 | F-type conjugal transfer protein TrbB | traF |
| KI226_RS22490 (ENKO_44020) | 78858..79127 | + | 270 | WP_254915067 | hypothetical protein | - |
| KI226_RS22495 (ENKO_44030) | 79120..80484 | + | 1365 | WP_088222287 | conjugal transfer pilus assembly protein TraH | traH |
| KI226_RS22500 (ENKO_44040) | 80524..83736 | + | 3213 | WP_088222286 | conjugal transfer mating-pair stabilization protein TraG | traG |
| KI226_RS22505 (ENKO_44050) | 83749..84321 | + | 573 | WP_088222285 | hypothetical protein | - |
| KI226_RS22510 (ENKO_44060) | 84466..85197 | + | 732 | WP_088222284 | complement resistance protein TraT | - |
| KI226_RS22515 (ENKO_44070) | 85413..85784 | + | 372 | WP_088222283 | hypothetical protein | - |
| KI226_RS22520 (ENKO_44080) | 85932..88427 | + | 2496 | WP_088222282 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 17143 | GenBank | NZ_AP024591 |
| Plasmid name | pENKO-1 | Incompatibility group | - |
| Plasmid size | 114856 bp | Coordinate of oriT [Strand] | 63314..63368 [-] |
| Host baterium | Enterobacter kobei strain JCM 8580 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | pcoE, pcoS, pcoR, pcoD, pcoC, pcoB, pcoA, silP, silA, silB, silF, silC, silR, silS, silE, arsC, arsB, arsR, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |