Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116626 |
| Name | oriT_CSUSB2|unnamed1 |
| Organism | Aeromonas hydrophila strain CSUSB2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP083945 (17285..17383 [-], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..30, 36..42 (TAACCGG..CCGGTTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_CSUSB2|unnamed1
GAATAAGGGACAGTGAAGATAGTTAACCGGCTTGCCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTGGGTACTCCAAGGAAAGTTTAC
GAATAAGGGACAGTGAAGATAGTTAACCGGCTTGCCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTGGGTACTCCAAGGAAAGTTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file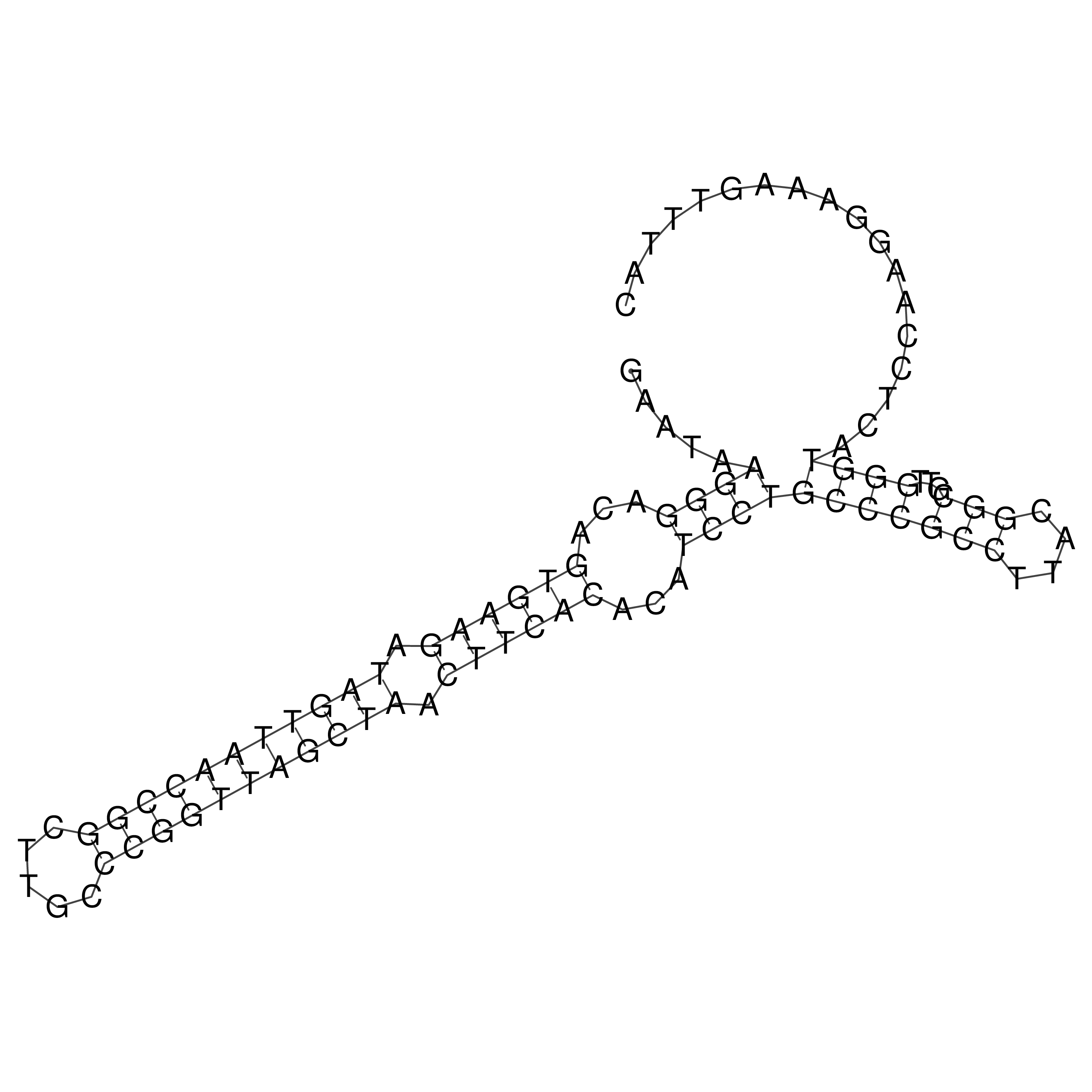
Relaxase
| ID | 10596 | GenBank | WP_224481346 |
| Name | traI_LCH17_RS22420_CSUSB2|unnamed1 |
UniProt ID | _ |
| Length | 950 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 950 a.a. Molecular weight: 104796.77 Da Isoelectric Point: 10.4940
>WP_224481346.1 TraI/MobA(P) family conjugative relaxase [Aeromonas hydrophila]
MIAKHVPMRSLGKSNFADLANYLTDEQSKEHRLGAVKVTNCESANLKAAIEEILATQHINTRAKSDKTYH
LIVSFRAGEEISDDVLKAIEERICAGLGFSDHQRVSAVHTDTDNLHIHIAINKIHPEKGTIHEPYYPHRT
LSELCELCERDYGLERDNHMPNKRGAAGRAQDMERHAGVESLVGWIKRECLTEIRAATTWEQLHKVLSDN
GLTLRERGNGLVFESSDGTAVKGSTLSRDLSKPALEAKLGAFISSSGEGKSEAKRTYDKKPVRSRVNTTE
LYARYRKEQKNLTSDRAEALERARRNKDKAVENAKRANSLRRATIKVVDSKGINKKVLYAQAFSVYRAKL
NTIHQEYAKERELLFKGFQRRTWADWLKHEAQNGNQQALNALRARDSAKGLQGNTITGQGTIIYRAGQSA
VRDDGDRLQVSANADREALTAALKIATERYAGRITVNGTPEFKAQMIRAAVDSQLPITFADPGLELRRQE
LLNKKEQDDARRREPDDRGRADRRSDGRDGARTTAHSRDDGGGNNPRPAGRERDTTRDNRKPNVGRIGRV
PPPESKNRLRALSKLGVVRLARGGEVLLSGHVPGHMEHQRSKPDHELRRGVSGAGGVTPPGYSPSSIIDN
ITKKGTIIYRAGQSAVRDDGDRLQVSTTTTPEVLSAALDMAAKRYGSCITVNGTPEFKAQMIRAAVDSQL
PITFADPGLELRRQELLNKKEQEDARRREQHDRGRADRRSDGRDGARTTAYGRNDGGGNPPRPAGRERSA
ASVDTTRDNRKPNVGRIGRVPPPESKNRLRALSKLGVVRIASGGEVLLPGHVPGHMEHQGSKPDHALQRG
VSGAGGVTQSETKAFGAVDKYIAEREAKRLLGFDISKHCRYNAEGGTLTFAGVRRIEDQALALLKRSDED
TVFVMPIDTATANRLKKISIGDAVSVTPKGSIKTSKGRSR
MIAKHVPMRSLGKSNFADLANYLTDEQSKEHRLGAVKVTNCESANLKAAIEEILATQHINTRAKSDKTYH
LIVSFRAGEEISDDVLKAIEERICAGLGFSDHQRVSAVHTDTDNLHIHIAINKIHPEKGTIHEPYYPHRT
LSELCELCERDYGLERDNHMPNKRGAAGRAQDMERHAGVESLVGWIKRECLTEIRAATTWEQLHKVLSDN
GLTLRERGNGLVFESSDGTAVKGSTLSRDLSKPALEAKLGAFISSSGEGKSEAKRTYDKKPVRSRVNTTE
LYARYRKEQKNLTSDRAEALERARRNKDKAVENAKRANSLRRATIKVVDSKGINKKVLYAQAFSVYRAKL
NTIHQEYAKERELLFKGFQRRTWADWLKHEAQNGNQQALNALRARDSAKGLQGNTITGQGTIIYRAGQSA
VRDDGDRLQVSANADREALTAALKIATERYAGRITVNGTPEFKAQMIRAAVDSQLPITFADPGLELRRQE
LLNKKEQDDARRREPDDRGRADRRSDGRDGARTTAHSRDDGGGNNPRPAGRERDTTRDNRKPNVGRIGRV
PPPESKNRLRALSKLGVVRLARGGEVLLSGHVPGHMEHQRSKPDHELRRGVSGAGGVTPPGYSPSSIIDN
ITKKGTIIYRAGQSAVRDDGDRLQVSTTTTPEVLSAALDMAAKRYGSCITVNGTPEFKAQMIRAAVDSQL
PITFADPGLELRRQELLNKKEQEDARRREQHDRGRADRRSDGRDGARTTAYGRNDGGGNPPRPAGRERSA
ASVDTTRDNRKPNVGRIGRVPPPESKNRLRALSKLGVVRIASGGEVLLPGHVPGHMEHQGSKPDHALQRG
VSGAGGVTQSETKAFGAVDKYIAEREAKRLLGFDISKHCRYNAEGGTLTFAGVRRIEDQALALLKRSDED
TVFVMPIDTATANRLKKISIGDAVSVTPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12380 | GenBank | WP_158114458 |
| Name | t4cp2_LCH17_RS22425_CSUSB2|unnamed1 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69901.89 Da Isoelectric Point: 7.5599
>WP_158114458.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Aeromonas]
MKIKMNNAVGPQVRDKAAKKSKVLPILGALSMGAGLQAATQYFAYAFNYQDVLGHNFNGIYAPWSILQWA
GQWYSDYPDEITRAGGAGILVSSVGLLAVAIGKVVAANTSKANEYLHGSARWAEKKDIQTAGLLPRDRNF
LEIVTGKDAPTATGVYVGGWEDKEGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASAVITDLK
GELWALTAGWRKTHARNKVLRFEPASSNGGVCWNPLDEIRVGTEYEVGDVQNLATLIVDPDGKGMESHWQ
KTAFSLLVGVILHALYKAKNEGTPATLPSIDAMMADPNRDIGELWMEMATYGHIDGQNHMAVGTAARDMM
DRPEEEAGSVLSTAKSYLSLYRDPVVARNVSKSEFRIRDLMHADDPASLYIVTQPNDKARLRPLVRIMAN
MIVRLLADKMDFENGRPVAHYKNRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDEQITSNCHVQNAYPPNRVETAEHLSRLTGQTTIVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGFPAIYGKQPLYFKDPVFQARASIAPPKVSDRFTTFPADGE
GIKI
MKIKMNNAVGPQVRDKAAKKSKVLPILGALSMGAGLQAATQYFAYAFNYQDVLGHNFNGIYAPWSILQWA
GQWYSDYPDEITRAGGAGILVSSVGLLAVAIGKVVAANTSKANEYLHGSARWAEKKDIQTAGLLPRDRNF
LEIVTGKDAPTATGVYVGGWEDKEGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASAVITDLK
GELWALTAGWRKTHARNKVLRFEPASSNGGVCWNPLDEIRVGTEYEVGDVQNLATLIVDPDGKGMESHWQ
KTAFSLLVGVILHALYKAKNEGTPATLPSIDAMMADPNRDIGELWMEMATYGHIDGQNHMAVGTAARDMM
DRPEEEAGSVLSTAKSYLSLYRDPVVARNVSKSEFRIRDLMHADDPASLYIVTQPNDKARLRPLVRIMAN
MIVRLLADKMDFENGRPVAHYKNRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDEQITSNCHVQNAYPPNRVETAEHLSRLTGQTTIVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGFPAIYGKQPLYFKDPVFQARASIAPPKVSDRFTTFPADGE
GIKI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 12381 | GenBank | WP_042655544 |
| Name | t4cp2_LCH17_RS22610_CSUSB2|unnamed1 |
UniProt ID | _ |
| Length | 845 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 845 a.a. Molecular weight: 94134.94 Da Isoelectric Point: 5.8295
>WP_042655544.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Aeromonas]
MIQGITISIAVLGAVLLLILFARIRAVDAELQLKKHRSKDAGLADLLIHAAVVDDGVIVGKNGSFMASWL
YKGNDNASSTDEQREMVSFRINQALAGLGNGWMVHIDAVRRPAPNYSEKGASNFPDPVSEALDEERRRLF
ESLGTMYEGYFVFTVTWFPPVLAQRKFVELMFDDDAVTPDHKARTKGLITQFKREISSIENRLSSAVEMI
RLKGQKIVCEDDTVVTHDDFLSWLQFCITGNHHPVILPSNPMYLDAIIGGQEMWGGVVPKIGRKFIQVVA
IEGFPLESTPGILAALAELPIEYRWSSRFIFMDQHESIAHLDKFRKKWRQKVRGFFDQVFNTNTGAIDQD
ALSMVQDAESAIAEINSGLIAMGYYTSLVVLMDEDRDALESSARSVEKAINRLGFAARIETINTLDAYLG
SIPGHGVENVRRPLINTMNLADLLPTSSIWTGSASAPCPLYPPQSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYKGMSIYAFDKGMSMYALTKATGGRHFTVAADDGKLAFCPLQFLET
KSDRAWAMEWIDTILALNGVETTPGQRNDIGHAIISMRDSGARTLSEFYTTIQDEQIREAIKQYTIDGAM
GHLLDAEEDGLQLSDFTTFEIEELMNLGEKYALPVLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVMRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNIYARDEDTSALYRRMGLNARQIE
IIATAVPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVATIQQLEAKFGDDWIHEWLSEKGLNIND
YLEAA
MIQGITISIAVLGAVLLLILFARIRAVDAELQLKKHRSKDAGLADLLIHAAVVDDGVIVGKNGSFMASWL
YKGNDNASSTDEQREMVSFRINQALAGLGNGWMVHIDAVRRPAPNYSEKGASNFPDPVSEALDEERRRLF
ESLGTMYEGYFVFTVTWFPPVLAQRKFVELMFDDDAVTPDHKARTKGLITQFKREISSIENRLSSAVEMI
RLKGQKIVCEDDTVVTHDDFLSWLQFCITGNHHPVILPSNPMYLDAIIGGQEMWGGVVPKIGRKFIQVVA
IEGFPLESTPGILAALAELPIEYRWSSRFIFMDQHESIAHLDKFRKKWRQKVRGFFDQVFNTNTGAIDQD
ALSMVQDAESAIAEINSGLIAMGYYTSLVVLMDEDRDALESSARSVEKAINRLGFAARIETINTLDAYLG
SIPGHGVENVRRPLINTMNLADLLPTSSIWTGSASAPCPLYPPQSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYKGMSIYAFDKGMSMYALTKATGGRHFTVAADDGKLAFCPLQFLET
KSDRAWAMEWIDTILALNGVETTPGQRNDIGHAIISMRDSGARTLSEFYTTIQDEQIREAIKQYTIDGAM
GHLLDAEEDGLQLSDFTTFEIEELMNLGEKYALPVLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVMRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNIYARDEDTSALYRRMGLNARQIE
IIATAVPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVATIQQLEAKFGDDWIHEWLSEKGLNIND
YLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46047..64676
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LCH17_RS22510 (LCH17_22510) | 42455..42943 | - | 489 | WP_157416887 | hypothetical protein | - |
| LCH17_RS22515 (LCH17_22515) | 42985..43905 | - | 921 | WP_082034755 | type II secretion system protein | - |
| LCH17_RS22520 (LCH17_22520) | 43908..44417 | - | 510 | WP_082034756 | type II secretion system protein | - |
| LCH17_RS22525 (LCH17_22525) | 44477..44986 | - | 510 | WP_050070469 | type II secretion system protein | - |
| LCH17_RS22530 (LCH17_22530) | 45013..46050 | - | 1038 | WP_042655532 | type II secretion system F family protein | - |
| LCH17_RS22535 (LCH17_22535) | 46047..47564 | - | 1518 | WP_042655533 | GspE/PulE family protein | virB11 |
| LCH17_RS22540 (LCH17_22540) | 47575..48255 | - | 681 | WP_042655534 | hypothetical protein | - |
| LCH17_RS22545 (LCH17_22545) | 48252..49394 | - | 1143 | WP_224481332 | hypothetical protein | - |
| LCH17_RS22550 (LCH17_22550) | 49397..50899 | - | 1503 | WP_042655536 | hypothetical protein | - |
| LCH17_RS22555 (LCH17_22555) | 50899..52185 | - | 1287 | WP_082034757 | OmpA family protein | - |
| LCH17_RS22560 (LCH17_22560) | 52398..53096 | - | 699 | WP_050070470 | TraX family protein | - |
| LCH17_RS22565 (LCH17_22565) | 53128..53766 | - | 639 | WP_042655553 | muramidase | - |
| LCH17_RS22570 (LCH17_22570) | 53779..54354 | - | 576 | WP_082034759 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| LCH17_RS22575 (LCH17_22575) | 54371..55942 | - | 1572 | WP_224481357 | P-type conjugative transfer protein TrbL | virB6 |
| LCH17_RS22580 (LCH17_22580) | 55947..56153 | - | 207 | WP_082034760 | entry exclusion lipoprotein TrbK | - |
| LCH17_RS22585 (LCH17_22585) | 56165..56932 | - | 768 | WP_042655540 | P-type conjugative transfer protein TrbJ | virB5 |
| LCH17_RS22590 (LCH17_22590) | 56953..58341 | - | 1389 | WP_042655541 | TrbI/VirB10 family protein | virB10 |
| LCH17_RS22595 (LCH17_22595) | 58345..58782 | - | 438 | WP_042655542 | conjugal transfer protein TrbH | - |
| LCH17_RS22600 (LCH17_22600) | 58785..59672 | - | 888 | WP_010676134 | P-type conjugative transfer protein TrbG | virB9 |
| LCH17_RS22605 (LCH17_22605) | 59690..60451 | - | 762 | WP_042655543 | conjugal transfer protein TrbF | virB8 |
| LCH17_RS22610 (LCH17_22610) | 60448..62985 | - | 2538 | WP_042655544 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| LCH17_RS22615 (LCH17_22615) | 62982..63293 | - | 312 | WP_010676131 | conjugal transfer protein TrbD | virB3 |
| LCH17_RS22620 (LCH17_22620) | 63297..63695 | - | 399 | WP_010676130 | conjugal transfer system pilin TrbC | virB2 |
| LCH17_RS22625 (LCH17_22625) | 63717..64676 | - | 960 | WP_042655545 | P-type conjugative transfer ATPase TrbB | virB11 |
| LCH17_RS22630 (LCH17_22630) | 64903..65298 | - | 396 | WP_042655546 | transcriptional regulator | - |
| LCH17_RS22635 (LCH17_22635) | 65404..65748 | + | 345 | WP_042655547 | single-stranded DNA-binding protein | - |
| LCH17_RS22640 (LCH17_22640) | 65927..66856 | + | 930 | WP_042655554 | replication protein RepA | - |
| LCH17_RS22645 (LCH17_22645) | 67637..68392 | - | 756 | WP_224481333 | tyrosine-type recombinase/integrase | - |
Host bacterium
| ID | 17059 | GenBank | NZ_CP083945 |
| Plasmid name | CSUSB2|unnamed1 | Incompatibility group | - |
| Plasmid size | 68825 bp | Coordinate of oriT [Strand] | 17285..17383 [-] |
| Host baterium | Aeromonas hydrophila strain CSUSB2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |