Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116610 |
| Name | oriT_pKA36387-KPC2 |
| Organism | Klebsiella aerogenes strain NRZ-36387 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP104947 (133470..133515 [+], 46 nt) |
| oriT length | 46 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 46 nt
>oriT_pKA36387-KPC2
AATCTGCAAAATTTTAATTTTGCGTGGTGTGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGTGTGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file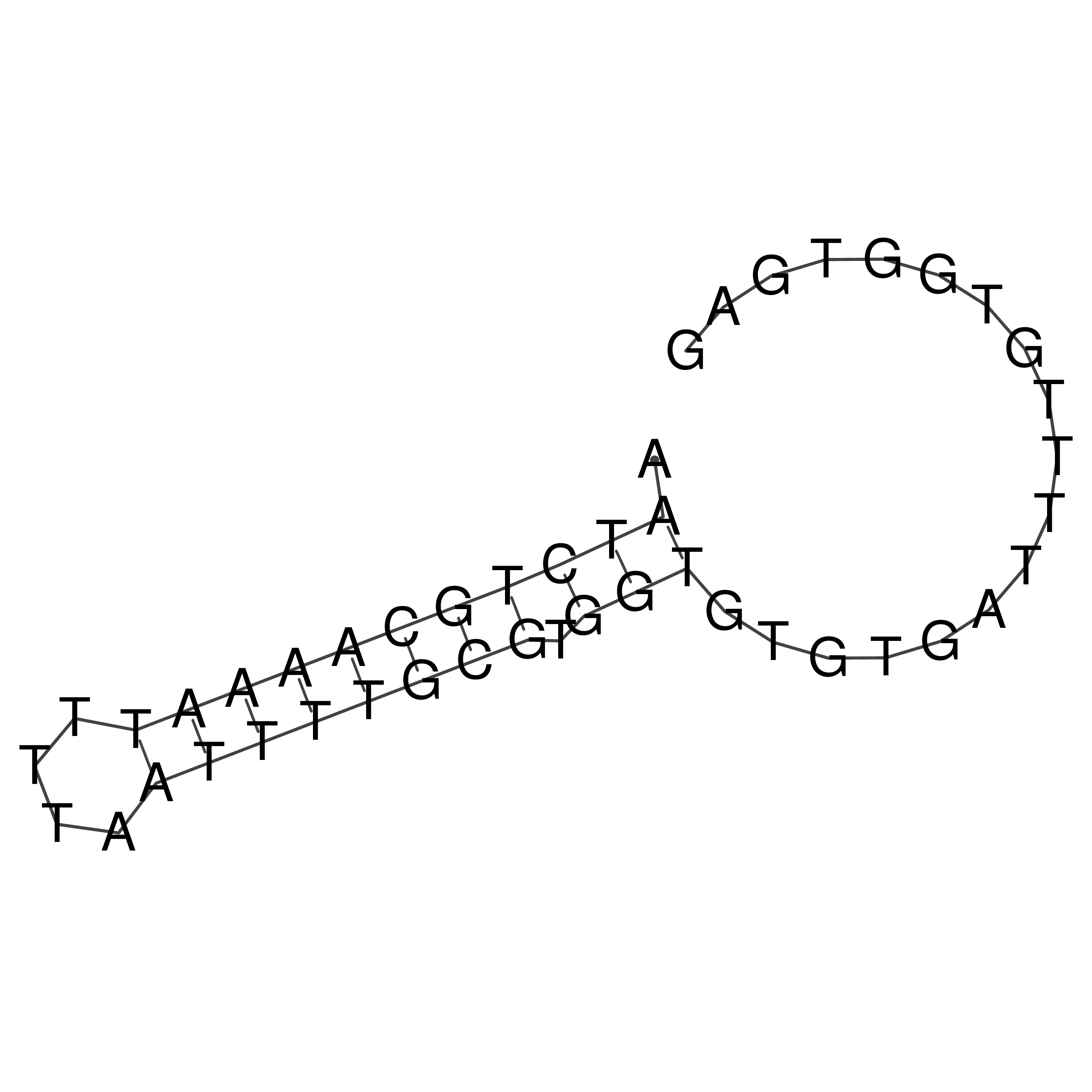
Relaxase
| ID | 10586 | GenBank | WP_001568074 |
| Name | traI_KKS27_RS00530_pKA36387-KPC2 |
UniProt ID | A0A9Q9P2W5 |
| Length | 1751 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1751 a.a. Molecular weight: 191075.29 Da Isoelectric Point: 6.1600
>WP_001568074.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGKAGDYYTKEDNYYVIGSMDERWQGKGAEALGLEGKTVDKALFTELLKGKLPDGSDLSRI
QDGMNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIEAHNQAVTEAIRQVETLAATRVMVDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREVLKPLVE
GLGYETEVVGKHGMWEMKDVPVEPFSTRTQEINAAAGPDASLKSRDVATLDTRKAKETLDPAQKVTEWLS
TLKETGFNLKEYREAADQRAKTQREVPSPEAPAGPNVDDIVTQAISGLSDRKVQFTYTELLARTVSQMEA
RPGVIETARAGIEAAIGREQLIPLDREKGIFTSGIHVLDELSVRALSQEVLRQNHVSMVPEKATARQTSF
SDAVTVLAQDRPAIGIVSGQGGAAGQRERVTEMAMMAREQGRDVHIIAADNRSLEFLLKEEKLAGEIITG
KSALQDGTAFVPGGTLIIDQAEKLSLKETINLLDGAMRNNVQILMSDSGKRSGTGSALTVMKESGVTAYK
WQGGKQTTASVISEPDRNQRFTRMASDFVASVRDGQESVVQIRGPREQGILTGLIRENLRNEGLIGSHEE
TITALTPVWLDSKNRGIRDNYREGMVMERWDPEKRIHDRFIIDRVTAVSNTLTLKDEKGESLNLKVSAVD
NTWTLFRPQQLPVAEGERLAILGKIPETRLKGGDTVQVLKAENGVLTVQHPGQKTTQKLAAGNTPFTGIK
VGHGWVENPGRSVSETATVFASLSQRELDNATLNALAQSGSQVRLYSAQNEQKTVEKLGRHTAFSVVTEQ
LKTRAGEQDISAAIAHQKSQLHTPAEQAIHLAVPSLESDKLTFTRPQLLVAAHEFSDGTVPFSDIEKTIG
EQIKSGDLLHVHVAAGHGNDLLISRQTWDAEKSLLTMVLEGKNAVTPLMAKVPGTLMEGLTSGQRNATRM
ILETRDRFTLVQGYAGVGKTTQFRAVMSAVNLLPEDSRPKVIGLGPTHRAVGEMQSAGVEAQTTASFLHD
TQLLMRSGQTPDFSNTLFLVDESSMLGLSDTAKAMSLIAAGGGRAVSSGDTDQLQSIAPGQPFRLMQKRS
AADVAIMQEIVRQVPELRPAVYSLIDRDIHSALSTIERVEPAQVPRKENAWVPESSVMEFTPEQEKAIQK
AIGEGEVLPSISPVSLYDAIVRDYTGRTESAQAQTLIITHLNDDRRVLNRMIHDEKMRNGELGEDSVTLP
VLVTSNIRDGELRKVSTWEAHKGAIALVDNQYHQIVGVSKADQLITLRSDEGTERLISPREASAEGVTLY
RQESITVSPGDRMRFSKSDTERGYVANSVWRVESISGDSVTLNDGKSTRTVTPAADKAEAHVDLAYAITT
HGAQGASEPFSIELEGVTGARERMVSFESAYVALSRMKQHTQVYTDDRKGWTDAVSRSQEKASAHDVLEP
LNDRAVKSAEQMFGRAIPLTESPAGRAALQQSGIARGQSTGKFISPGKKYPLPHVALPAFDRNGKSAGIW
LSPLTDKDGRLQAIGGEGRVMGNEEARFVALQGSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPGAITGGRVWGDIDAVVTGGSGANIPLPPDLLAQKTAEELQRKQIEKQAQETARELTGEKRKQDEVGE
QIKDIITDVVRGLERQKGQHERFQLPDEPENRLRNEAVQKVVADNQQQDRLQQIERDMVRDLTREKTLGG
D
MMSIGSVKSAGKAGDYYTKEDNYYVIGSMDERWQGKGAEALGLEGKTVDKALFTELLKGKLPDGSDLSRI
QDGMNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIEAHNQAVTEAIRQVETLAATRVMVDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREVLKPLVE
GLGYETEVVGKHGMWEMKDVPVEPFSTRTQEINAAAGPDASLKSRDVATLDTRKAKETLDPAQKVTEWLS
TLKETGFNLKEYREAADQRAKTQREVPSPEAPAGPNVDDIVTQAISGLSDRKVQFTYTELLARTVSQMEA
RPGVIETARAGIEAAIGREQLIPLDREKGIFTSGIHVLDELSVRALSQEVLRQNHVSMVPEKATARQTSF
SDAVTVLAQDRPAIGIVSGQGGAAGQRERVTEMAMMAREQGRDVHIIAADNRSLEFLLKEEKLAGEIITG
KSALQDGTAFVPGGTLIIDQAEKLSLKETINLLDGAMRNNVQILMSDSGKRSGTGSALTVMKESGVTAYK
WQGGKQTTASVISEPDRNQRFTRMASDFVASVRDGQESVVQIRGPREQGILTGLIRENLRNEGLIGSHEE
TITALTPVWLDSKNRGIRDNYREGMVMERWDPEKRIHDRFIIDRVTAVSNTLTLKDEKGESLNLKVSAVD
NTWTLFRPQQLPVAEGERLAILGKIPETRLKGGDTVQVLKAENGVLTVQHPGQKTTQKLAAGNTPFTGIK
VGHGWVENPGRSVSETATVFASLSQRELDNATLNALAQSGSQVRLYSAQNEQKTVEKLGRHTAFSVVTEQ
LKTRAGEQDISAAIAHQKSQLHTPAEQAIHLAVPSLESDKLTFTRPQLLVAAHEFSDGTVPFSDIEKTIG
EQIKSGDLLHVHVAAGHGNDLLISRQTWDAEKSLLTMVLEGKNAVTPLMAKVPGTLMEGLTSGQRNATRM
ILETRDRFTLVQGYAGVGKTTQFRAVMSAVNLLPEDSRPKVIGLGPTHRAVGEMQSAGVEAQTTASFLHD
TQLLMRSGQTPDFSNTLFLVDESSMLGLSDTAKAMSLIAAGGGRAVSSGDTDQLQSIAPGQPFRLMQKRS
AADVAIMQEIVRQVPELRPAVYSLIDRDIHSALSTIERVEPAQVPRKENAWVPESSVMEFTPEQEKAIQK
AIGEGEVLPSISPVSLYDAIVRDYTGRTESAQAQTLIITHLNDDRRVLNRMIHDEKMRNGELGEDSVTLP
VLVTSNIRDGELRKVSTWEAHKGAIALVDNQYHQIVGVSKADQLITLRSDEGTERLISPREASAEGVTLY
RQESITVSPGDRMRFSKSDTERGYVANSVWRVESISGDSVTLNDGKSTRTVTPAADKAEAHVDLAYAITT
HGAQGASEPFSIELEGVTGARERMVSFESAYVALSRMKQHTQVYTDDRKGWTDAVSRSQEKASAHDVLEP
LNDRAVKSAEQMFGRAIPLTESPAGRAALQQSGIARGQSTGKFISPGKKYPLPHVALPAFDRNGKSAGIW
LSPLTDKDGRLQAIGGEGRVMGNEEARFVALQGSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPGAITGGRVWGDIDAVVTGGSGANIPLPPDLLAQKTAEELQRKQIEKQAQETARELTGEKRKQDEVGE
QIKDIITDVVRGLERQKGQHERFQLPDEPENRLRNEAVQKVVADNQQQDRLQQIERDMVRDLTREKTLGG
D
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6213 | GenBank | WP_263433060 |
| Name | WP_263433060_pKA36387-KPC2 |
UniProt ID | _ |
| Length | 128 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 128 a.a. Molecular weight: 14939.04 Da Isoelectric Point: 5.0657
>WP_263433060.1 conjugal transfer relaxosome DNA-binding protein TraM [Klebsiella aerogenes]
MARVQAYPSDEVVDKINAIVEKRRAEVRRKKDISFSSVATMLLELGLRVYEAQMERKESGFNQKEFNKVL
LENVMKTQFTISKVLAMDSLSPHINGDDRFDFKSMVMNIRDDAREVVERFFPSQDEEE
MARVQAYPSDEVVDKINAIVEKRRAEVRRKKDISFSSVATMLLELGLRVYEAQMERKESGFNQKEFNKVL
LENVMKTQFTISKVLAMDSLSPHINGDDRFDFKSMVMNIRDDAREVVERFFPSQDEEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12369 | GenBank | WP_001568075 |
| Name | traD_KKS27_RS00535_pKA36387-KPC2 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 82704.57 Da Isoelectric Point: 5.9222
>WP_001568075.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIASMRFRMFGQIANIILYVLFLFFWVLCGLILMYRLSWQTFVNGAVYWWCTTLGPMR
DIIRSQSVYTINYYGQQLQYTSEQILKDKYTIWCGEQLWTGFVFAGTVSLIICIVAFFVASWVLGHQGKQ
QSEDEVTGGRQLSEKPKEVARKMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSIDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRSVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKGIA
EFAAGEIGEKEIKKASENYSYGADPVRDGVSTGKEQKRETIVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRQLNQAIDDKLAAVLAAREAEGQSARILFMPDPVETVPVEKTDEKPASPIAAVPTP
QEDKAKVPPVTASNTFHKPASAAAAAASASVTQAGGVEQELHEKPEEQLPLPPGVNKDGEIEDMNAWDEW
QSSSDVLRDMHRREEVNINHSHHVDRDDINLGSNF
MSFNAKDMTQGGQIASMRFRMFGQIANIILYVLFLFFWVLCGLILMYRLSWQTFVNGAVYWWCTTLGPMR
DIIRSQSVYTINYYGQQLQYTSEQILKDKYTIWCGEQLWTGFVFAGTVSLIICIVAFFVASWVLGHQGKQ
QSEDEVTGGRQLSEKPKEVARKMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSIDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRSVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKGIA
EFAAGEIGEKEIKKASENYSYGADPVRDGVSTGKEQKRETIVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRQLNQAIDDKLAAVLAAREAEGQSARILFMPDPVETVPVEKTDEKPASPIAAVPTP
QEDKAKVPPVTASNTFHKPASAAAAAASASVTQAGGVEQELHEKPEEQLPLPPGVNKDGEIEDMNAWDEW
QSSSDVLRDMHRREEVNINHSHHVDRDDINLGSNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 75034..85359
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KKS27_RS00325 (KKS27_p00330) | 70810..71790 | + | 981 | WP_000019473 | IS5-like element ISKpn26 family transposase | - |
| KKS27_RS00330 (KKS27_p00335) | 72128..73009 | + | 882 | WP_004199234 | carbapenem-hydrolyzing class A beta-lactamase KPC-2 | - |
| KKS27_RS00335 (KKS27_p00340) | 73259..74578 | - | 1320 | WP_004152397 | IS1182-like element ISKpn6 family transposase | - |
| KKS27_RS00340 (KKS27_p00345) | 74930..75034 | - | 105 | Protein_70 | phospholipase D family protein | - |
| KKS27_RS00345 (KKS27_p00350) | 75034..76029 | - | 996 | WP_012561144 | ATPase, T2SS/T4P/T4SS family | virB11 |
| KKS27_RS00350 (KKS27_p00355) | 76071..77231 | - | 1161 | WP_000101710 | type IV secretion system protein VirB10 | virB10 |
| KKS27_RS00355 (KKS27_p00360) | 77231..78115 | - | 885 | WP_000735066 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| KKS27_RS00360 (KKS27_p00365) | 78126..78824 | - | 699 | WP_000646594 | virB8 family protein | virB8 |
| KKS27_RS00365 (KKS27_p00375) | 79043..79546 | - | 504 | WP_263433057 | type IV secretion system protein | virB6 |
| KKS27_RS00370 (KKS27_p00380) | 79558..79752 | - | 195 | Protein_76 | hypothetical protein | - |
| KKS27_RS00375 (KKS27_p00385) | 79673..80080 | - | 408 | Protein_77 | type IV secretion system protein | - |
| KKS27_RS00380 (KKS27_p00390) | 80096..80322 | - | 227 | Protein_78 | IncN-type entry exclusion lipoprotein EexN | - |
| KKS27_RS00385 (KKS27_p00395) | 80330..81043 | - | 714 | WP_001749960 | type IV secretion system protein | virB5 |
| KKS27_RS00390 (KKS27_p00400) | 81061..83661 | - | 2601 | WP_012561149 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| KKS27_RS00395 (KKS27_p00405) | 83661..83978 | - | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| KKS27_RS00400 (KKS27_p00410) | 84029..84322 | - | 294 | WP_001749962 | hypothetical protein | virB2 |
| KKS27_RS00405 (KKS27_p00415) | 84332..84613 | - | 282 | WP_016338364 | transcriptional repressor KorA | - |
| KKS27_RS00410 (KKS27_p00420) | 84622..85359 | - | 738 | WP_013279384 | lytic transglycosylase domain-containing protein | virB1 |
| KKS27_RS00415 (KKS27_p00425) | 85468..85773 | + | 306 | WP_012561154 | H-NS family nucleoid-associated regulatory protein | - |
| KKS27_RS00420 (KKS27_p00430) | 85789..86133 | + | 345 | WP_012561155 | hypothetical protein | - |
| KKS27_RS00425 (KKS27_p00435) | 86130..86444 | + | 315 | WP_016338363 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| KKS27_RS00430 (KKS27_p00440) | 86480..86791 | + | 312 | WP_013279382 | hypothetical protein | - |
| KKS27_RS00435 (KKS27_p00445) | 86847..87107 | + | 261 | WP_016359294 | hypothetical protein | - |
| KKS27_RS00440 (KKS27_p00450) | 87148..88116 | - | 969 | WP_016151349 | IS5 family transposase | - |
| KKS27_RS00445 (KKS27_p00460) | 88200..88553 | + | 354 | WP_225622145 | restriction endonuclease | - |
| KKS27_RS00450 (KKS27_p00465) | 88558..88764 | + | 207 | WP_001749967 | hypothetical protein | - |
| KKS27_RS00455 (KKS27_p00470) | 88775..89047 | - | 273 | Protein_93 | IS1 family transposase | - |
Region 2: 108469..134072
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KKS27_RS00535 (KKS27_p00550) | 108469..110676 | - | 2208 | WP_001568075 | type IV conjugative transfer system coupling protein TraD | virb4 |
| KKS27_RS00540 (KKS27_p00555) | 110861..111589 | - | 729 | Protein_110 | conjugal transfer complement resistance protein TraT | - |
| KKS27_RS28850 (KKS27_p00560) | 111767..112293 | - | 527 | Protein_111 | conjugal transfer protein TraS | - |
| KKS27_RS00545 (KKS27_p00565) | 112299..115148 | - | 2850 | WP_263433058 | conjugal transfer mating-pair stabilization protein TraG | traG |
| KKS27_RS00550 (KKS27_p00570) | 115148..116518 | - | 1371 | WP_023332869 | conjugal transfer pilus assembly protein TraH | traH |
| KKS27_RS00555 (KKS27_p00575) | 116505..117041 | - | 537 | WP_032160235 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| KKS27_RS00560 (KKS27_p00580) | 117061..117294 | - | 234 | WP_001568081 | type-F conjugative transfer system pilin chaperone TraQ | - |
| KKS27_RS00565 (KKS27_p00585) | 117305..118054 | - | 750 | WP_001568082 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| KKS27_RS00570 (KKS27_p00590) | 118070..118402 | - | 333 | WP_001568083 | hypothetical protein | - |
| KKS27_RS00575 (KKS27_p00595) | 118399..118659 | - | 261 | WP_001568084 | conjugal transfer protein TrbE | - |
| KKS27_RS00580 (KKS27_p00600) | 118672..120563 | - | 1892 | Protein_119 | type-F conjugative transfer system mating-pair stabilization protein TraN | - |
| KKS27_RS00585 (KKS27_p00605) | 120560..121186 | - | 627 | WP_213492003 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| KKS27_RS00590 (KKS27_p00610) | 121199..122191 | - | 993 | WP_023332868 | conjugal transfer pilus assembly protein TraU | traU |
| KKS27_RS00595 (KKS27_p00615) | 122202..122831 | - | 630 | WP_001568088 | type-F conjugative transfer system protein TraW | traW |
| KKS27_RS00600 (KKS27_p00620) | 122828..123205 | - | 378 | WP_001568089 | type-F conjugative transfer system protein TrbI | - |
| KKS27_RS00605 (KKS27_p00625) | 123205..125844 | - | 2640 | WP_001568090 | type IV secretion system protein TraC | virb4 |
| KKS27_RS00610 (KKS27_p00635) | 126305..126703 | - | 399 | WP_001568092 | hypothetical protein | - |
| KKS27_RS00615 (KKS27_p00640) | 126708..127121 | - | 414 | WP_001568093 | hypothetical protein | - |
| KKS27_RS00620 (KKS27_p00645) | 127235..127804 | - | 570 | WP_001568094 | type IV conjugative transfer system lipoprotein TraV | traV |
| KKS27_RS00625 (KKS27_p00650) | 128084..129463 | - | 1380 | Protein_128 | F-type conjugal transfer pilus assembly protein TraB | - |
| KKS27_RS00630 (KKS27_p00655) | 129463..130203 | - | 741 | WP_001568101 | type-F conjugative transfer system secretin TraK | traK |
| KKS27_RS00635 (KKS27_p00660) | 130190..130756 | - | 567 | WP_001568102 | type IV conjugative transfer system protein TraE | traE |
| KKS27_RS00640 (KKS27_p00665) | 130775..131080 | - | 306 | WP_024191986 | type IV conjugative transfer system protein TraL | traL |
| KKS27_RS00645 (KKS27_p00670) | 131094..131462 | - | 369 | WP_001568105 | type IV conjugative transfer system pilin TraA | - |
| KKS27_RS00650 (KKS27_p00675) | 131541..131705 | - | 165 | WP_071594636 | TraY domain-containing protein | - |
| KKS27_RS00655 (KKS27_p00680) | 132013..132387 | - | 375 | WP_263433059 | hypothetical protein | - |
| KKS27_RS00660 (KKS27_p00685) | 132783..133169 | - | 387 | WP_263433060 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| KKS27_RS00665 (KKS27_p00690) | 133587..134072 | + | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| KKS27_RS00670 (KKS27_p00695) | 134105..134430 | - | 326 | Protein_137 | DUF5983 family protein | - |
| KKS27_RS00675 (KKS27_p00700) | 134463..135283 | - | 821 | Protein_138 | DUF932 domain-containing protein | - |
| KKS27_RS00680 (KKS27_p00705) | 136089..136486 | - | 398 | Protein_139 | hypothetical protein | - |
| KKS27_RS00685 (KKS27_p00710) | 136662..137472 | - | 811 | Protein_140 | type III restriction endonuclease subunit R | - |
| KKS27_RS00690 (KKS27_p00715) | 137544..138520 | + | 977 | Protein_141 | IS5 family transposase | - |
| KKS27_RS28855 | 138519..138677 | + | 159 | Protein_142 | EAL domain-containing protein | - |
Host bacterium
| ID | 17043 | GenBank | NZ_CP104947 |
| Plasmid name | pKA36387-KPC2 | Incompatibility group | IncFIA |
| Plasmid size | 165682 bp | Coordinate of oriT [Strand] | 133470..133515 [+] |
| Host baterium | Klebsiella aerogenes strain NRZ-36387 |
Cargo genes
| Drug resistance gene | aph(6)-Id, aph(3'')-Ib, aac(3)-IId, blaTEM-1B, blaKPC-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |