Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116454 |
| Name | oriT_pNIID-071400-001-1 |
| Organism | Clostridium tetani strain NIID-071400-001 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP026824 (24568..24771 [-], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file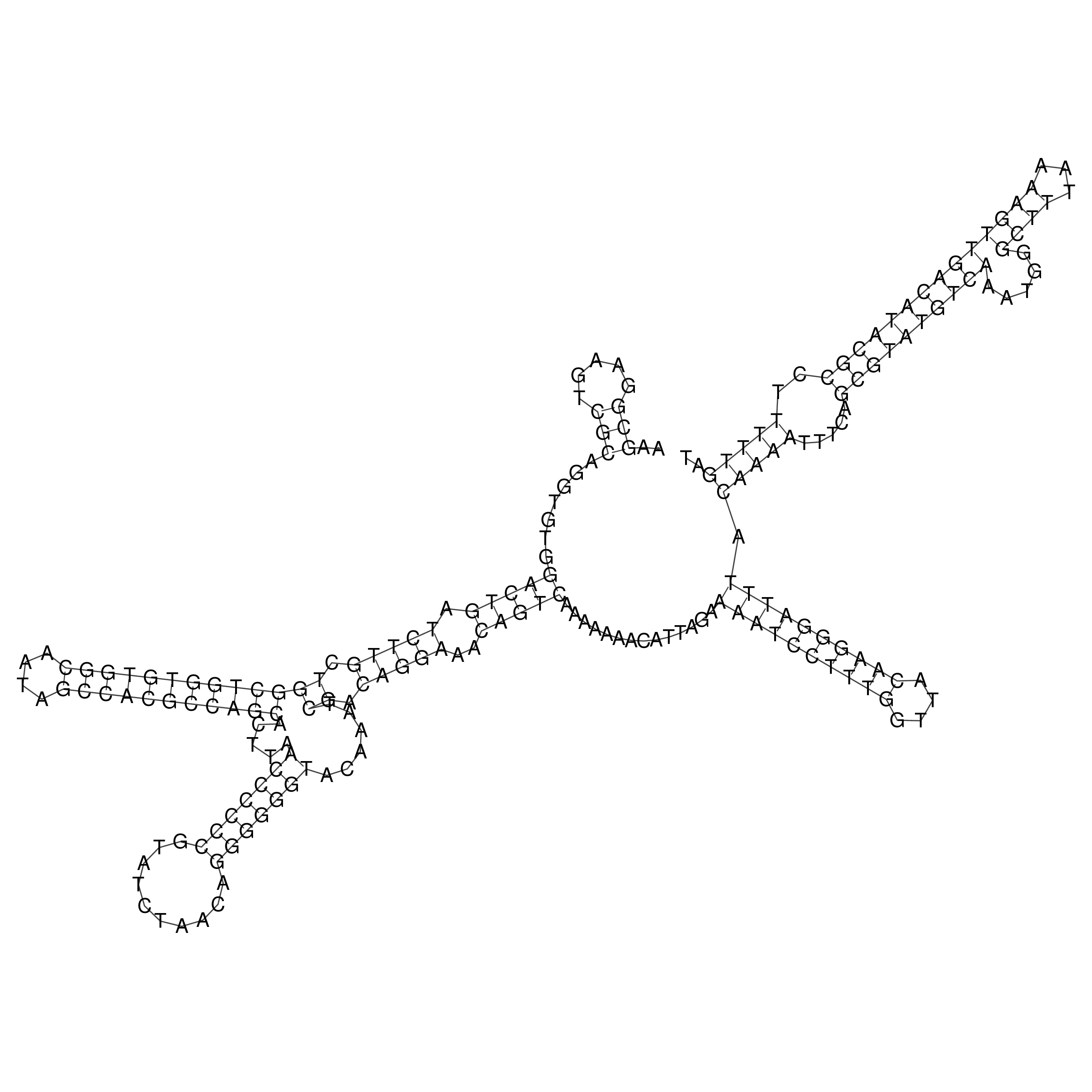
Relaxase
| ID | 10514 | GenBank | WP_000398284 |
| Name | mobT_R9A53_RS13890_pNIID-071400-001-1 |
UniProt ID | A0A3P3PY43 |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3PY43 |
Auxiliary protein
| ID | 6175 | GenBank | WP_001291561 |
| Name | WP_001291561_pNIID-071400-001-1 |
UniProt ID | A0A3P3Q021 |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3Q021 |
T4CP
| ID | 12279 | GenBank | WP_000813488 |
| Name | tcpA_R9A53_RS13900_pNIID-071400-001-1 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 12280 | GenBank | WP_035142135 |
| Name | t4cp2_R9A53_RS14395_pNIID-071400-001-1 |
UniProt ID | _ |
| Length | 776 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 776 a.a. Molecular weight: 89803.17 Da Isoelectric Point: 7.4131
MKSIKISKYFEFINPEYVYLKITPDKSIRNYNSSSIAKSIAHTYKSISQRIRKEQKKIFFETNFKISYII
DIYSNDVEFYFIVPNPFVKIITEKILEVWPNVTIKESNRVRAYSSSAISYQLKYKKEDALSLAVDKKTNE
PLNSILSVIDILDEGDRVTIVYNFLPRNKLGWTKQYNDTMKKIKEHKPLERQKLALEYIVKSVISLLIHL
IDSISEVLNDFAGGNNKNNENLLEAVITSLSNEKDISIATKKKKDMDILDTQIAILSSSNDEIRQTSNVL
SVCQAYRVLDQDNELLYSRVNNISNFKLDDYKLKGIETNTISTDEAQNFIQLPGRRLLSTFNINHINTTE
IQVPHDLQKGFFYLGEVKYKGIKTKAFLEDEYNIGNLPLVLIGSQGSGKTTYIKNIAKYCNKNKEGLFIL
DFIKNCEMSSDISKIIPEDDLVILNLSKEDDMQGLGYNEINITEDMTTFERLNLANLQSQQAMALIDSIS
IGDALSSRMRRFFNAAANVVFSQNYSSIKSVIECLESHIKRDMYINNLSDELKAMLEDEINTLKELDEYS
KASKGHPKIELIGTRESKIEHILDRVNMLREDFKLKYMYNKPTKNNINLVELMEEGKTVIIQMKESDFPT
KMIKNTLVTYWISKIWLTSQLRGSLHEKPLRCNVIVDEIFQAPTCMKTLEYILPQCRKFGCKFIFSTQYI
RQLDDIFDTLEASGSSYMLLKGCMEDDFNHLKSKLYNFEYEDLRDMKQWHSLNLVYYSKGYSSFISKLPS
PINDKQ
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 15466..26876
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| R9A53_RS13815 (N071400001_p10110) | 10653..10856 | - | 204 | WP_000814511 | excisionase | - |
| R9A53_RS13820 | 10840..11091 | + | 252 | WP_001845478 | hypothetical protein | - |
| R9A53_RS13825 (N071400001_p10120) | 11317..11547 | - | 231 | WP_000857133 | helix-turn-helix domain-containing protein | - |
| R9A53_RS13830 (N071400001_p10130) | 11544..11966 | - | 423 | WP_000804885 | sigma-70 family RNA polymerase sigma factor | - |
| R9A53_RS13835 (N071400001_p10140) | 12471..12824 | + | 354 | WP_001227347 | helix-turn-helix transcriptional regulator | - |
| R9A53_RS13840 | 12884..13051 | - | 168 | WP_000336323 | cysteine-rich KTR domain-containing protein | - |
| R9A53_RS13845 (N071400001_p10150) | 13170..15089 | - | 1920 | WP_317739008 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| R9A53_RS13850 | 15105..15221 | - | 117 | WP_001814923 | tetracycline resistance determinant leader peptide | - |
| R9A53_RS13855 (N071400001_p10160) | 15466..16398 | - | 933 | WP_001224318 | conjugal transfer protein | orf13 |
| R9A53_RS13860 (N071400001_p10170) | 16395..17396 | - | 1002 | WP_000769868 | lysozyme family protein | orf14 |
| R9A53_RS13865 (N071400001_p10180) | 17393..19570 | - | 2178 | WP_000804748 | CD3337/EF1877 family mobilome membrane protein | orf15 |
| R9A53_RS13870 (N071400001_p10190) | 19573..22020 | - | 2448 | WP_000331160 | ATP-binding protein | virb4 |
| R9A53_RS13875 | 22004..22396 | - | 393 | WP_000723888 | conjugal transfer protein | orf17a |
| R9A53_RS13880 (N071400001_p10200) | 22485..22982 | - | 498 | WP_000342539 | antirestriction protein ArdA | - |
| R9A53_RS13885 (N071400001_p10210) | 23099..23320 | - | 222 | WP_001009056 | hypothetical protein | orf19 |
| R9A53_RS13890 (N071400001_p10220) | 23363..24568 | - | 1206 | WP_000398284 | MobT family relaxase | - |
| R9A53_RS13895 | 24591..24743 | - | 153 | WP_000879507 | hypothetical protein | - |
| R9A53_RS13900 (N071400001_p10230) | 24746..26131 | - | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| R9A53_RS13905 (N071400001_p10240) | 26160..26546 | - | 387 | WP_000985015 | YdcP family protein | orf23 |
| R9A53_RS13910 (N071400001_p10250) | 26562..26876 | - | 315 | WP_000420682 | YdcP family protein | orf23 |
| R9A53_RS13915 (N071400001_p10260) | 27211..28281 | - | 1071 | WP_317739009 | ABC transporter permease | - |
| R9A53_RS13920 (N071400001_p10280) | 29356..30042 | - | 687 | WP_035141379 | N-acetylmuramoyl-L-alanine amidase | - |
| R9A53_RS13925 (N071400001_p10290) | 30415..30714 | - | 300 | WP_035141380 | hypothetical protein | - |
| R9A53_RS13930 (N071400001_p10300) | 31039..31257 | - | 219 | WP_035141381 | DUF2922 domain-containing protein | - |
| R9A53_RS13935 (N071400001_p10310) | 31281..31502 | - | 222 | WP_035109232 | DUF1659 domain-containing protein | - |
| R9A53_RS13940 (N071400001_p10320) | 31591..31770 | - | 180 | WP_035111455 | YvrJ family protein | - |
Host bacterium
| ID | 16887 | GenBank | NZ_AP026824 |
| Plasmid name | pNIID-071400-001-1 | Incompatibility group | - |
| Plasmid size | 112887 bp | Coordinate of oriT [Strand] | 24568..24771 [-] |
| Host baterium | Clostridium tetani strain NIID-071400-001 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | tetX |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |