Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116411 |
| Name | oriT_NML110041|unnamed1 |
| Organism | Burkholderia cenocepacia strain NML110041 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP102478 (75969..76669 [+], 701 nt) |
| oriT length | 701 nt |
| IRs (inverted repeats) | 597..602, 607..612 (GCGGGG..CCCCGC) 535..540, 545..550 (CGCGCC..GGCGCG) 175..183, 197..205 (TGTCAGACA..TGTCTGACA) 37..45, 51..59 (GAGTCGAGG..CCTCGACTC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 701 nt
>oriT_NML110041|unnamed1
GAAAAACGCCTGGAATGAAAAGAGCGGGCGCTCGTGGAGTCGAGGAAAAACCTCGACTCGCGCACGAAACCGCGCTCGATTACGGCCCTCATGACTGCATTATTTTGAGCGACAGTAGCATGGCGACGAGCGTTTCGCAATAACCCATTTATTTTCATAGTGATACTTATCGAATGTCAGACACACCCGTTCTAGCTGTCTGACAACGTGGCCCAATCATGACGGGGCGGGCGCGACATACTGGCAATGTTCGCGCTTTCCGGTCATTCGGGTGTCTGACATCGCGAGCCTTAACGCAAGCCGTTTCTCGCGATACGTGGCGGCGTTTTCTCGACAGGTCGACGGTGGCTGTCTGACGCTGCAAACCGCTACCGTCAGTGGTCGAGCGGGAATTTTCTATTCCTCTGGAGTCCGAATTATATTCTCCGATTCCGAATTTGTCTGACACCGGAGGATATGACGTACAAGGGGTCGCGCTTGATCCCTGCCTCGCTTTTTTCGCCTATTTTCGTAGGTGTCTGACGCCGAAGGCACCGCGCCATACGGCGCGACACACCGCAACAACGTTGCATGGTGTGTGACGATCGTCTTAGGGGGCGGGGATTCCCCCGCTTCCTAAGACGATCGTAGCCAACTTCACTTCAGGTTTGGGCCGGTCGCGGAGCCTTCACCCGGTAAGGTACGGCGGTTCCCGCTCAAGG
GAAAAACGCCTGGAATGAAAAGAGCGGGCGCTCGTGGAGTCGAGGAAAAACCTCGACTCGCGCACGAAACCGCGCTCGATTACGGCCCTCATGACTGCATTATTTTGAGCGACAGTAGCATGGCGACGAGCGTTTCGCAATAACCCATTTATTTTCATAGTGATACTTATCGAATGTCAGACACACCCGTTCTAGCTGTCTGACAACGTGGCCCAATCATGACGGGGCGGGCGCGACATACTGGCAATGTTCGCGCTTTCCGGTCATTCGGGTGTCTGACATCGCGAGCCTTAACGCAAGCCGTTTCTCGCGATACGTGGCGGCGTTTTCTCGACAGGTCGACGGTGGCTGTCTGACGCTGCAAACCGCTACCGTCAGTGGTCGAGCGGGAATTTTCTATTCCTCTGGAGTCCGAATTATATTCTCCGATTCCGAATTTGTCTGACACCGGAGGATATGACGTACAAGGGGTCGCGCTTGATCCCTGCCTCGCTTTTTTCGCCTATTTTCGTAGGTGTCTGACGCCGAAGGCACCGCGCCATACGGCGCGACACACCGCAACAACGTTGCATGGTGTGTGACGATCGTCTTAGGGGGCGGGGATTCCCCCGCTTCCTAAGACGATCGTAGCCAACTTCACTTCAGGTTTGGGCCGGTCGCGGAGCCTTCACCCGGTAAGGTACGGCGGTTCCCGCTCAAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file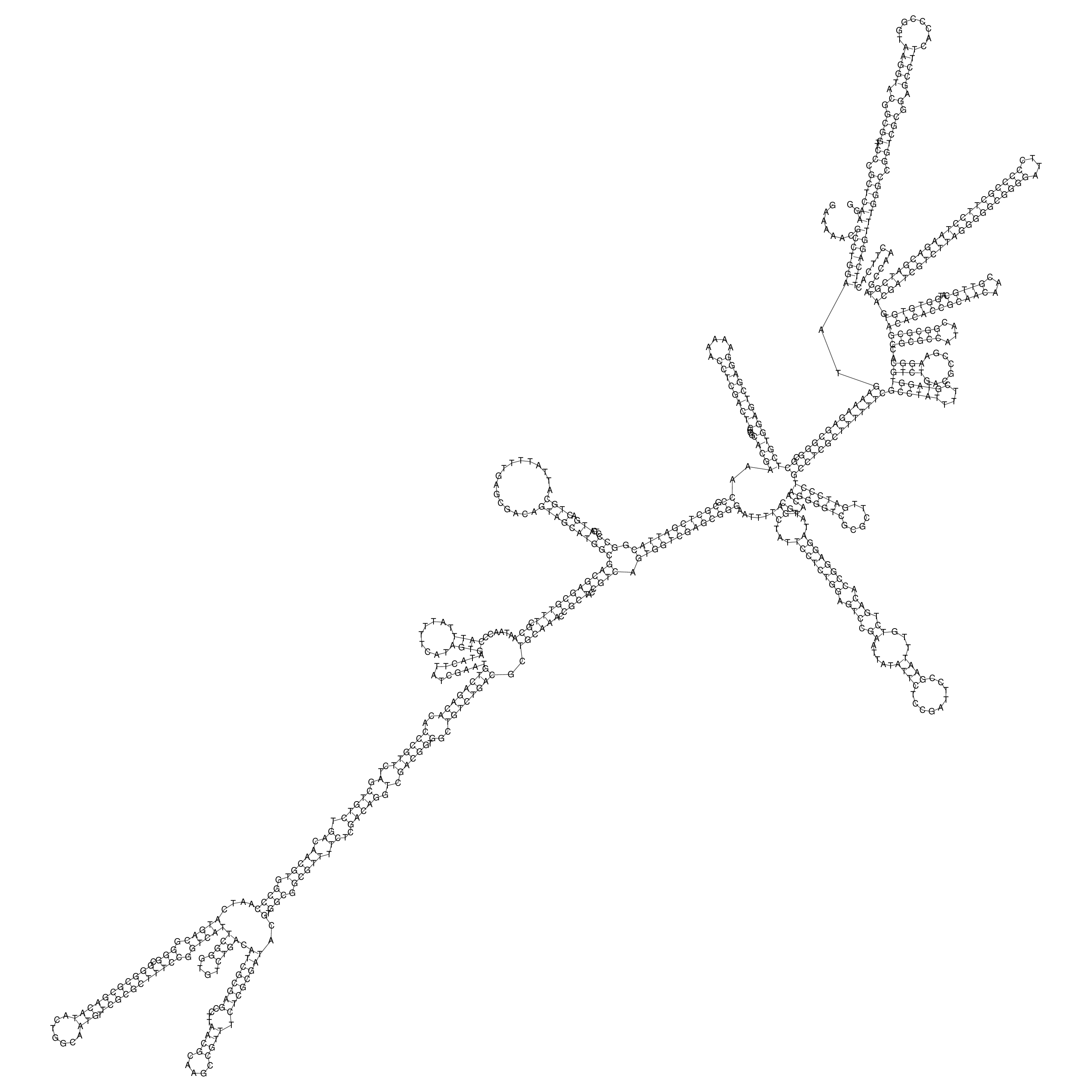
Relaxase
| ID | 10489 | GenBank | WP_006488612 |
| Name | mobF_NUJ27_RS36820_NML110041|unnamed1 |
UniProt ID | B4EQJ6 |
| Length | 939 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 939 a.a. Molecular weight: 104810.59 Da Isoelectric Point: 6.4316
>WP_006488612.1 MobF family relaxase [Burkholderia cenocepacia]
MLSLHNIASAAQTRNYFSKDNYYTQDENTKHSAWFGAGAARLGLAGQVQPAAFLNLLEGRVEEQQLGKWV
TDEETGERRQDHQPGIDITFSAPKSVSLMAEVAGDTAVRDAHEAAVQSALAYLEENVIRTRSTQNGVTSI
EHTGNMVAALFKHNTSRDLDPQTHTHAVVMNATLRADGKWRSIDNREIYQTQKLAGAIYNAELVSGLQQL
GYRLTNPDRHGNFEIQGISREQIEHFSQRRTAMKEKLGKRQLELDTASAADKERAALATRERKVDVDHEQ
LLADWQARALEQGIDLGALRATAAERKAEGIDPPTKLTGRQALAFAAAHLIEREAVVDKSDLVETAVVHG
TGRVSPQEVMKAFRQLEKNGDLVPLPDEKYTTRKMLESERWALDFVKQEKGNAPVVLEATKVQERIAEAE
RLQQFSYTFGQRDAIVLALTSKDRFVAIQGLAGTGKTTMLRTLNDIAAAEGYVVRGMAPTGAAAKVMVKE
TGIAAETVAMFQIKERQLQKDIEFAQQFAPDFKRKPEVWVVDESSFLAQRQMAQLHRLAEHAGAKVVYLG
DTLQLQGVEAGKPFELAQKDGIATAYMTDINRQKTDVMKEAVDIITGRNQLDAGTRLTDIELKRNGQSFQ
YLSQQGRVHETDEVVTSTVDKILAMSPEERARNITITPLNRDRVAINEGVRVGLQARGELSKNESTHEIL
VSKGFTRAQIKEAQYYERGDVVRFNRTYQQINADKGDYTRVIDVDHESGIVTLRKRDGSTLAWEPAKHNK
VEVYDPDLRQLAVGDVIRLTRNDETFKNGETARVVQLTGEHAVLQVDDRGVISHHSVNLETSQHWDHAYA
STVHAAQGASRNTTIFAINMPDNEDKKAQEREFQALAKVFGDRSYYVGVTRASHDISIVTNDIDLAQRAI
TGKLDKTTVIEAMNQEVREQTIQREQHEI
MLSLHNIASAAQTRNYFSKDNYYTQDENTKHSAWFGAGAARLGLAGQVQPAAFLNLLEGRVEEQQLGKWV
TDEETGERRQDHQPGIDITFSAPKSVSLMAEVAGDTAVRDAHEAAVQSALAYLEENVIRTRSTQNGVTSI
EHTGNMVAALFKHNTSRDLDPQTHTHAVVMNATLRADGKWRSIDNREIYQTQKLAGAIYNAELVSGLQQL
GYRLTNPDRHGNFEIQGISREQIEHFSQRRTAMKEKLGKRQLELDTASAADKERAALATRERKVDVDHEQ
LLADWQARALEQGIDLGALRATAAERKAEGIDPPTKLTGRQALAFAAAHLIEREAVVDKSDLVETAVVHG
TGRVSPQEVMKAFRQLEKNGDLVPLPDEKYTTRKMLESERWALDFVKQEKGNAPVVLEATKVQERIAEAE
RLQQFSYTFGQRDAIVLALTSKDRFVAIQGLAGTGKTTMLRTLNDIAAAEGYVVRGMAPTGAAAKVMVKE
TGIAAETVAMFQIKERQLQKDIEFAQQFAPDFKRKPEVWVVDESSFLAQRQMAQLHRLAEHAGAKVVYLG
DTLQLQGVEAGKPFELAQKDGIATAYMTDINRQKTDVMKEAVDIITGRNQLDAGTRLTDIELKRNGQSFQ
YLSQQGRVHETDEVVTSTVDKILAMSPEERARNITITPLNRDRVAINEGVRVGLQARGELSKNESTHEIL
VSKGFTRAQIKEAQYYERGDVVRFNRTYQQINADKGDYTRVIDVDHESGIVTLRKRDGSTLAWEPAKHNK
VEVYDPDLRQLAVGDVIRLTRNDETFKNGETARVVQLTGEHAVLQVDDRGVISHHSVNLETSQHWDHAYA
STVHAAQGASRNTTIFAINMPDNEDKKAQEREFQALAKVFGDRSYYVGVTRASHDISIVTNDIDLAQRAI
TGKLDKTTVIEAMNQEVREQTIQREQHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B4EQJ6 |
T4CP
| ID | 12242 | GenBank | WP_006488610 |
| Name | t4cp2_NUJ27_RS36830_NML110041|unnamed1 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 72007.72 Da Isoelectric Point: 6.9695
>WP_006488610.1 type IV secretion system DNA-binding domain-containing protein [Burkholderia cenocepacia]
MTGKKPNGLFQDFSTGSEIWTHRVALVLVAIRNILLLSLFLGLAAGFIYAAFFVDRDIWRPAVANVIAHL
RSVVMMTAVPMEYEVNGQTVRLPVDQVVALTDPVWDVAYFFLRRFLIIVATVTACTCVSIAAYWYEFGRS
TMADKDLRGATLVNGKQLAKMIRQDDMESPYSIAGVPMLRNAETLHTLVVGAQGTGKSQQFFDRLRQVRA
RGKRAIVYDPSGEFAEEFFREGKDVMMNPFDDRSPNWNIKYEIEEEYHYDSMATALIPDPREADPFWSLA
GREVFRDVARTLHREGNLTNRNLYNSIAMSNLDAIYQLLRNTAGASYVDPKTERTGMSLKMTVQNQLSSF
RYLRDDGKPFSIRQWIYNESDSWMFISTREEMLEAIKPIMSLWIDIAIKAVLNLTPIHRERLHFSIDEVP
TLQKLDIMKLAVSNTRKYGLCMMLGAQDMPQFLDIYGEYLARTIINGCQTKLLLRVTDAEAAEMFSKIIG
QTEVEEKDESLSFGVNSQRDGFSIAKRRLLRDLALPSQILRLRDMTGYLVYPGEYPIARVKYGFVKPRKN
APAFIKRQAFSELFAPAAGSPATAAASESVVAGPSPDWIAHPEILVAASGGVDPETGEILPPRPGAWRWG
DGNQRGDHA
MTGKKPNGLFQDFSTGSEIWTHRVALVLVAIRNILLLSLFLGLAAGFIYAAFFVDRDIWRPAVANVIAHL
RSVVMMTAVPMEYEVNGQTVRLPVDQVVALTDPVWDVAYFFLRRFLIIVATVTACTCVSIAAYWYEFGRS
TMADKDLRGATLVNGKQLAKMIRQDDMESPYSIAGVPMLRNAETLHTLVVGAQGTGKSQQFFDRLRQVRA
RGKRAIVYDPSGEFAEEFFREGKDVMMNPFDDRSPNWNIKYEIEEEYHYDSMATALIPDPREADPFWSLA
GREVFRDVARTLHREGNLTNRNLYNSIAMSNLDAIYQLLRNTAGASYVDPKTERTGMSLKMTVQNQLSSF
RYLRDDGKPFSIRQWIYNESDSWMFISTREEMLEAIKPIMSLWIDIAIKAVLNLTPIHRERLHFSIDEVP
TLQKLDIMKLAVSNTRKYGLCMMLGAQDMPQFLDIYGEYLARTIINGCQTKLLLRVTDAEAAEMFSKIIG
QTEVEEKDESLSFGVNSQRDGFSIAKRRLLRDLALPSQILRLRDMTGYLVYPGEYPIARVKYGFVKPRKN
APAFIKRQAFSELFAPAAGSPATAAASESVVAGPSPDWIAHPEILVAASGGVDPETGEILPPRPGAWRWG
DGNQRGDHA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 34802..63117
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NUJ27_RS36580 (NUJ27_36580) | 30053..30625 | - | 573 | WP_006487431 | PH domain-containing protein | - |
| NUJ27_RS36585 (NUJ27_36585) | 30622..30789 | - | 168 | WP_006487400 | hypothetical protein | - |
| NUJ27_RS36590 (NUJ27_36590) | 30786..32102 | - | 1317 | WP_006487416 | DUF1173 domain-containing protein | - |
| NUJ27_RS36595 (NUJ27_36595) | 32099..33511 | - | 1413 | WP_006487358 | zeta toxin family protein | - |
| NUJ27_RS36600 (NUJ27_36600) | 33522..33788 | - | 267 | WP_006487406 | hypothetical protein | - |
| NUJ27_RS36605 (NUJ27_36605) | 34432..34812 | + | 381 | WP_020980049 | hypothetical protein | - |
| NUJ27_RS36610 (NUJ27_36610) | 34802..37840 | - | 3039 | WP_006487373 | conjugal transfer protein TraG N-terminal domain-containing protein | traG |
| NUJ27_RS36615 (NUJ27_36615) | 37850..39304 | - | 1455 | WP_006487442 | conjugal transfer protein TraH | traH |
| NUJ27_RS36620 (NUJ27_36620) | 39297..40331 | - | 1035 | WP_006487369 | phage Gp37/Gp68 family protein | - |
| NUJ27_RS36625 (NUJ27_36625) | 40328..40771 | - | 444 | WP_006487377 | HNH endonuclease | - |
| NUJ27_RS36630 (NUJ27_36630) | 40768..41298 | - | 531 | WP_006487388 | hypothetical protein | - |
| NUJ27_RS36635 (NUJ27_36635) | 41309..42142 | - | 834 | WP_006487410 | conjugal transfer protein TraF | traF |
| NUJ27_RS36640 (NUJ27_36640) | 42147..42536 | - | 390 | WP_006493947 | hypothetical protein | - |
| NUJ27_RS36645 (NUJ27_36645) | 42533..42841 | - | 309 | WP_006493941 | hypothetical protein | - |
| NUJ27_RS36650 (NUJ27_36650) | 42838..43104 | - | 267 | WP_174900806 | hypothetical protein | - |
| NUJ27_RS36655 (NUJ27_36655) | 43101..44435 | - | 1335 | WP_263059962 | conjugal transfer protein TraN | traN |
| NUJ27_RS36660 (NUJ27_36660) | 44428..45245 | + | 818 | WP_085964347 | IS5-like element IS402 family transposase | - |
| NUJ27_RS36665 (NUJ27_36665) | 45270..46391 | - | 1122 | WP_006487394 | hypothetical protein | - |
| NUJ27_RS36670 (NUJ27_36670) | 46393..46770 | - | 378 | WP_006493944 | hypothetical protein | - |
| NUJ27_RS36675 (NUJ27_36675) | 46772..47473 | - | 702 | WP_006487447 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| NUJ27_RS36680 (NUJ27_36680) | 47470..48480 | - | 1011 | WP_006487375 | conjugal transfer pilus assembly protein TraU | traU |
| NUJ27_RS36685 (NUJ27_36685) | 48477..49115 | - | 639 | WP_006487379 | type-F conjugative transfer system protein TraW | traW |
| NUJ27_RS36690 (NUJ27_36690) | 49102..49632 | - | 531 | WP_006487449 | signal peptidase I | - |
| NUJ27_RS36695 (NUJ27_36695) | 49650..49868 | - | 219 | WP_006487354 | hypothetical protein | - |
| NUJ27_RS36700 (NUJ27_36700) | 49846..50658 | - | 813 | WP_006487420 | FadR/GntR family transcriptional regulator | - |
| NUJ27_RS36705 (NUJ27_36705) | 50672..51205 | - | 534 | WP_020980048 | phospholipase D family protein | - |
| NUJ27_RS36710 (NUJ27_36710) | 51281..51646 | - | 366 | WP_006487376 | hypothetical protein | - |
| NUJ27_RS36715 (NUJ27_36715) | 51643..52035 | - | 393 | WP_006493936 | hypothetical protein | - |
| NUJ27_RS36720 (NUJ27_36720) | 52032..52646 | - | 615 | WP_006487383 | hypothetical protein | - |
| NUJ27_RS36725 (NUJ27_36725) | 52643..53383 | - | 741 | WP_006487402 | hypothetical protein | - |
| NUJ27_RS36730 (NUJ27_36730) | 53380..53895 | - | 516 | WP_006487408 | hypothetical protein | - |
| NUJ27_RS36735 (NUJ27_36735) | 53892..56498 | - | 2607 | WP_006487362 | type IV secretion system protein TraC | virb4 |
| NUJ27_RS36740 (NUJ27_36740) | 56501..57163 | - | 663 | WP_006487356 | type IV conjugative transfer system lipoprotein TraV | traV |
| NUJ27_RS36745 (NUJ27_36745) | 57160..57954 | - | 795 | WP_006487398 | DsbC family protein | trbB |
| NUJ27_RS36750 (NUJ27_36750) | 57947..59200 | - | 1254 | WP_263059960 | TraB/VirB10 family protein | traB |
| NUJ27_RS36760 (NUJ27_36760) | 60290..61036 | - | 747 | WP_012493842 | type-F conjugative transfer system secretin TraK | traK |
| NUJ27_RS36765 (NUJ27_36765) | 60975..61565 | - | 591 | WP_006487381 | TraE/TraK family type IV conjugative transfer system protein | traE |
| NUJ27_RS36770 (NUJ27_36770) | 61572..61865 | - | 294 | WP_006487365 | type IV conjugative transfer system protein TraL | traL |
| NUJ27_RS36775 (NUJ27_36775) | 61879..62271 | - | 393 | WP_006487426 | hypothetical protein | - |
| NUJ27_RS36780 (NUJ27_36780) | 62300..62644 | - | 345 | WP_006493942 | hypothetical protein | - |
| NUJ27_RS36785 (NUJ27_36785) | 62641..63117 | - | 477 | WP_006487418 | lytic transglycosylase domain-containing protein | virB1 |
| NUJ27_RS36790 (NUJ27_36790) | 63552..63824 | - | 273 | WP_006493934 | hypothetical protein | - |
| NUJ27_RS36795 (NUJ27_36795) | 63946..64716 | - | 771 | WP_263059961 | 5-oxoprolinase subunit PxpA | - |
| NUJ27_RS36800 (NUJ27_36800) | 64703..64939 | - | 237 | WP_006487429 | hypothetical protein | - |
| NUJ27_RS36805 (NUJ27_36805) | 64936..65976 | - | 1041 | WP_006493948 | TRAP transporter substrate-binding protein DctP | - |
| NUJ27_RS36810 (NUJ27_36810) | 66099..66959 | - | 861 | WP_227743146 | LuxR family transcriptional regulator | - |
Host bacterium
| ID | 16844 | GenBank | NZ_CP102478 |
| Plasmid name | NML110041|unnamed1 | Incompatibility group | - |
| Plasmid size | 94494 bp | Coordinate of oriT [Strand] | 75969..76669 [+] |
| Host baterium | Burkholderia cenocepacia strain NML110041 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |