Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116405 |
| Name | oriT_FDAARGOS_80|unnamed1 |
| Organism | Proteus mirabilis strain FDAARGOS_80 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP026060 (1707..1807 [-], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 32..38, 43..49 (TTACGAT..ATCGTAA) 21..26, 38..43 (TTTTCA..TGAAAA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_FDAARGOS_80|unnamed1
AATTAGAATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATACCATACCACGCCTTTTTTAAGG
AATTAGAATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATACCATACCACGCCTTTTTTAAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file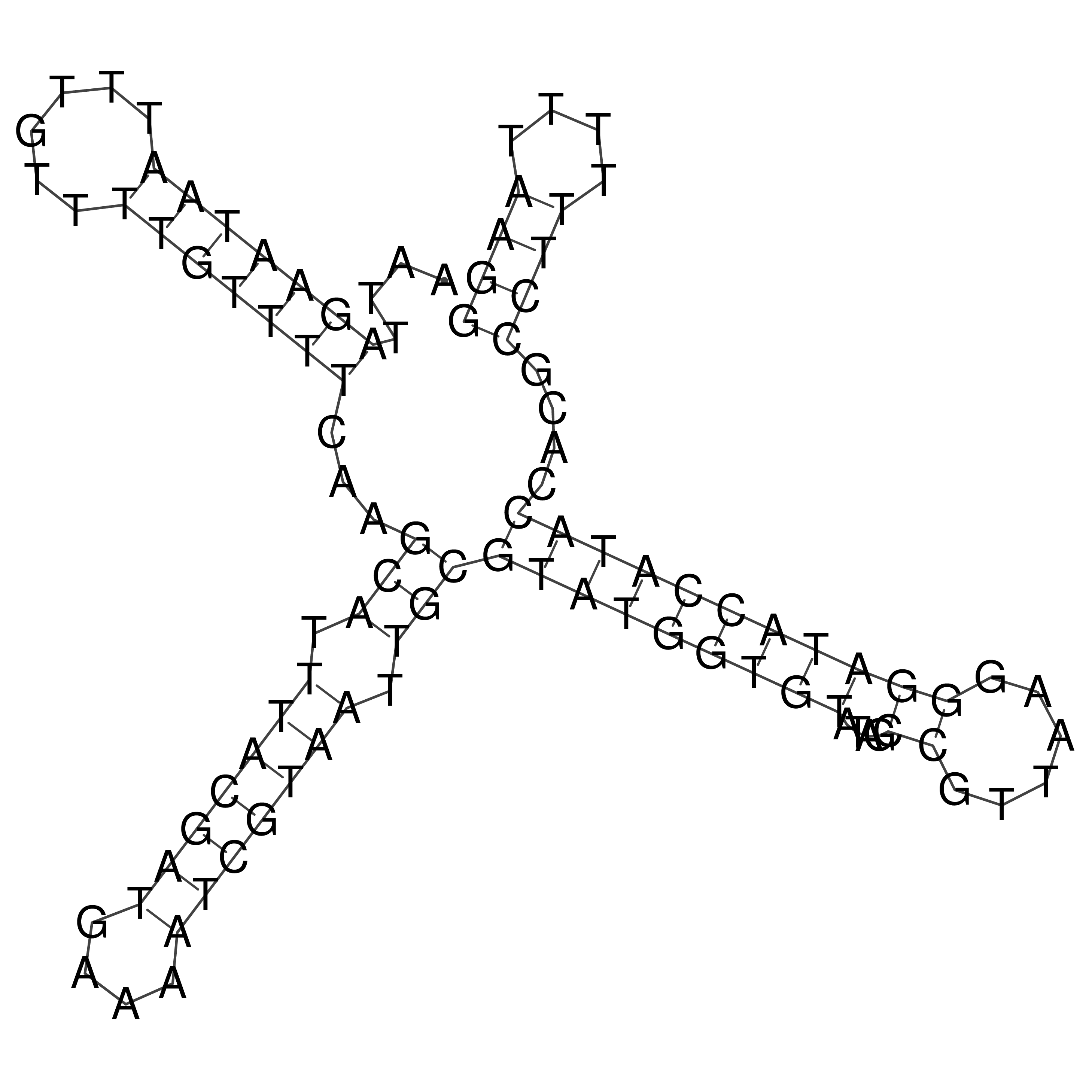
Relaxase
| ID | 10484 | GenBank | WP_000932317 |
| Name | mobF_MC72_RS18410_FDAARGOS_80|unnamed1 |
UniProt ID | A0A626R2M1 |
| Length | 969 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 969 a.a. Molecular weight: 108425.51 Da Isoelectric Point: 9.7925
>WP_000932317.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLNITTITRQRVDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISVM
RKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVENT
NKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGYN
LRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYEG
WKNRAKELQIDFDSREWKGAANDDIARNTQPAALEKPIEYQADKVMTFAIRSLTERQSVITERELMDVAM
KHGYGRLTLDDIRAARERAITSGNLIKEEATYAAATTNKKQQNVAMTREQWVTELVKAGRNKDEARKLVD
KGITNGRLVQQKPRFTTQLAQQRERNILKMEREGRGKIQTPYTREFSEGWLASRTLKPEQLKAVMGIIHT
PNQFISVHGFAGTGKSYMTKSAADFLKEQGVHVTSLAPYGSQVKALQAEGLESRTLQSFLRASDKKIGPG
SVVFIDEAGVIPARQMEETMRVIRDAGARAVFLGDTKQTKAVEAGKPFEQLIKAGMETAYMKDIQRQKDP
ELLKAALNAAEGKIKASLTHVTSIVEEKNHDQRYRRIVGDYVAMPPTDRANALIITGTNDSRKKINAYIQ
AELGLKGQGINYPLLNRLDTTQAQRQHSKYYEKGAVIIPERDYSNGLKRGAVYTVLDTGPGNKLTVSDGS
GDTIAFSPARFSKLSVYSVEKTELAVGDQVRITRNDAHLDLANGDRFKVKGVQSGEVLLENEKGRLVKID
ANKPMYLGLAYASTVHSAQGLTCDKVFINMDTRSLTTAKDVYYVAVTRAKHEAVIYTDDEKKLDKAASRE
GFKTAALELEQLKRYAKEQQHDAREKGRDKTEPTQEKDNAKRKGKGHDKNNDERAFTSL
MLNITTITRQRVDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISVM
RKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVENT
NKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGYN
LRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYEG
WKNRAKELQIDFDSREWKGAANDDIARNTQPAALEKPIEYQADKVMTFAIRSLTERQSVITERELMDVAM
KHGYGRLTLDDIRAARERAITSGNLIKEEATYAAATTNKKQQNVAMTREQWVTELVKAGRNKDEARKLVD
KGITNGRLVQQKPRFTTQLAQQRERNILKMEREGRGKIQTPYTREFSEGWLASRTLKPEQLKAVMGIIHT
PNQFISVHGFAGTGKSYMTKSAADFLKEQGVHVTSLAPYGSQVKALQAEGLESRTLQSFLRASDKKIGPG
SVVFIDEAGVIPARQMEETMRVIRDAGARAVFLGDTKQTKAVEAGKPFEQLIKAGMETAYMKDIQRQKDP
ELLKAALNAAEGKIKASLTHVTSIVEEKNHDQRYRRIVGDYVAMPPTDRANALIITGTNDSRKKINAYIQ
AELGLKGQGINYPLLNRLDTTQAQRQHSKYYEKGAVIIPERDYSNGLKRGAVYTVLDTGPGNKLTVSDGS
GDTIAFSPARFSKLSVYSVEKTELAVGDQVRITRNDAHLDLANGDRFKVKGVQSGEVLLENEKGRLVKID
ANKPMYLGLAYASTVHSAQGLTCDKVFINMDTRSLTTAKDVYYVAVTRAKHEAVIYTDDEKKLDKAASRE
GFKTAALELEQLKRYAKEQQHDAREKGRDKTEPTQEKDNAKRKGKGHDKNNDERAFTSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A626R2M1 |
T4CP
| ID | 12236 | GenBank | WP_036896339 |
| Name | t4cp2_MC72_RS18405_FDAARGOS_80|unnamed1 |
UniProt ID | _ |
| Length | 525 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 525 a.a. Molecular weight: 59001.90 Da Isoelectric Point: 9.5507
>WP_036896339.1 type IV secretion system DNA-binding domain-containing protein [Proteus mirabilis]
MNEADKGKICLGALLFTPLVTWFIAAKTLYGFTPKDARERLVLLIQQTFDLWPLWGALLVGEIIAIIGLI
IFFKFNHQIFKGAPFKKIYRGTRLVSQRALASLTKEREPQITIADIPIPTKAEGTHISIAGATGVGKSTI
FAEMMKGCLIRGKMQRVASKKDRMIILDPDGDFLSRFYKKGDKILNPYDARTEGWVFFNEIKSDFDFERF
GVSIVPTAKDSQTEEWNGYGRLLFTEVSRKVFNTSRNPTMEEVFYWTNEVPLEELEEYVRGTKAQALFAG
SGRAVGSARFVLSNKLAAHLKMPAGNFSIREWLEDPKGGNLYITWTDEMREALKSLISCWTDSIFSIVLG
MSKSNTRKIWTYIDELESLDYLPSLRDALTKGRKKGLRVVTGYQSYSQVISIYGSDVAETLLSNHRTSVV
MAAGRLGERTLEFVSKSLGEIEGEREKTGISRRFGQMGTKNINDDHKRERAVTPTEIATLDDLTGYIAFP
GNLPVAKFETHHVNYTRSNPVPGVVLRESTAFAGM
MNEADKGKICLGALLFTPLVTWFIAAKTLYGFTPKDARERLVLLIQQTFDLWPLWGALLVGEIIAIIGLI
IFFKFNHQIFKGAPFKKIYRGTRLVSQRALASLTKEREPQITIADIPIPTKAEGTHISIAGATGVGKSTI
FAEMMKGCLIRGKMQRVASKKDRMIILDPDGDFLSRFYKKGDKILNPYDARTEGWVFFNEIKSDFDFERF
GVSIVPTAKDSQTEEWNGYGRLLFTEVSRKVFNTSRNPTMEEVFYWTNEVPLEELEEYVRGTKAQALFAG
SGRAVGSARFVLSNKLAAHLKMPAGNFSIREWLEDPKGGNLYITWTDEMREALKSLISCWTDSIFSIVLG
MSKSNTRKIWTYIDELESLDYLPSLRDALTKGRKKGLRVVTGYQSYSQVISIYGSDVAETLLSNHRTSVV
MAAGRLGERTLEFVSKSLGEIEGEREKTGISRRFGQMGTKNINDDHKRERAVTPTEIATLDDLTGYIAFP
GNLPVAKFETHHVNYTRSNPVPGVVLRESTAFAGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2249..20038
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MC72_RS18390 (MC72_018390) | 311..1021 | - | 711 | WP_000686338 | StbB family protein | - |
| MC72_RS18395 (MC72_018395) | 1034..1477 | - | 444 | WP_024249956 | hypothetical protein | - |
| MC72_RS18400 (MC72_018400) | 1909..2256 | + | 348 | WP_001139329 | hypothetical protein | - |
| MC72_RS18405 (MC72_018405) | 2249..3826 | + | 1578 | WP_036896339 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| MC72_RS18410 (MC72_018410) | 3829..6738 | + | 2910 | WP_000932317 | MobF family relaxase | - |
| MC72_RS18415 (MC72_018415) | 7016..7210 | - | 195 | WP_000529710 | hypothetical protein | - |
| MC72_RS18420 (MC72_018420) | 7222..7704 | - | 483 | WP_036896345 | GNAT family N-acetyltransferase | - |
| MC72_RS18425 (MC72_018425) | 7717..8271 | - | 555 | WP_000616063 | phospholipase D family protein | - |
| MC72_RS18430 (MC72_018430) | 8234..9265 | - | 1032 | WP_000138004 | P-type DNA transfer ATPase VirB11 | virB11 |
| MC72_RS18435 (MC72_018435) | 9258..10412 | - | 1155 | WP_001224296 | type IV secretion system protein VirB10 | virB10 |
| MC72_RS18440 (MC72_018440) | 10409..11296 | - | 888 | WP_000744355 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| MC72_RS18445 (MC72_018445) | 11299..12015 | - | 717 | WP_000914645 | virB8 family protein | virB8 |
| MC72_RS19120 | 12018..12188 | - | 171 | WP_021536809 | hypothetical protein | - |
| MC72_RS18450 (MC72_018450) | 12283..13326 | - | 1044 | WP_000885742 | type IV secretion system protein | virB6 |
| MC72_RS18455 (MC72_018455) | 13345..13581 | - | 237 | WP_000733633 | EexN family lipoprotein | - |
| MC72_RS18460 (MC72_018460) | 13585..14292 | - | 708 | WP_036896352 | type IV secretion system protein | virB5 |
| MC72_RS18465 (MC72_018465) | 14302..16932 | - | 2631 | WP_000257330 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| MC72_RS18470 (MC72_018470) | 16932..17246 | - | 315 | WP_001053888 | type IV secretion system protein VirB3 | virB3 |
| MC72_RS18475 (MC72_018475) | 17296..17586 | - | 291 | WP_001090269 | TrbC/VirB2 family protein | virB2 |
| MC72_RS18480 (MC72_018480) | 17570..17887 | - | 318 | WP_001267114 | hypothetical protein | - |
| MC72_RS18485 (MC72_018485) | 17910..18650 | - | 741 | WP_000815895 | lytic transglycosylase domain-containing protein | virB1 |
| MC72_RS18490 (MC72_018490) | 18713..18949 | + | 237 | WP_000089855 | H-NS family nucleoid-associated regulatory protein | - |
| MC72_RS18495 (MC72_018495) | 18961..19398 | + | 438 | WP_000034608 | hypothetical protein | - |
| MC72_RS18500 (MC72_018500) | 19404..19721 | + | 318 | WP_001226231 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| MC72_RS18505 (MC72_018505) | 19724..20038 | + | 315 | WP_000851068 | hypothetical protein | tfc15 |
| MC72_RS18510 (MC72_018510) | 20014..20739 | + | 726 | WP_223307254 | restriction endonuclease | - |
| MC72_RS19460 | 20740..21141 | + | 402 | WP_023187076 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| MC72_RS18520 (MC72_018520) | 21201..21515 | - | 315 | WP_001280992 | hypothetical protein | - |
| MC72_RS18525 (MC72_018525) | 21541..21867 | - | 327 | WP_000817647 | endoribonuclease MazF | - |
| MC72_RS18530 (MC72_018530) | 21857..22153 | - | 297 | WP_223307255 | hypothetical protein | - |
| MC72_RS18535 (MC72_018535) | 22123..22665 | - | 543 | WP_000711249 | hypothetical protein | - |
| MC72_RS18540 (MC72_018540) | 22665..22910 | - | 246 | WP_000195442 | AbrB/MazE/SpoVT family DNA-binding domain-containing protein | - |
| MC72_RS18550 (MC72_018550) | 23372..23794 | + | 423 | WP_000838908 | hypothetical protein | - |
| MC72_RS18555 (MC72_018555) | 23794..24378 | + | 585 | WP_000690619 | recombinase family protein | - |
Host bacterium
| ID | 16838 | GenBank | NZ_CP026060 |
| Plasmid name | FDAARGOS_80|unnamed1 | Incompatibility group | - |
| Plasmid size | 34615 bp | Coordinate of oriT [Strand] | 1707..1807 [-] |
| Host baterium | Proteus mirabilis strain FDAARGOS_80 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |