Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116364 |
| Name | oriT_pK118-KPC |
| Organism | Citrobacter koseri strain K118 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP136818 (76545..76593 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pK118-KPC
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file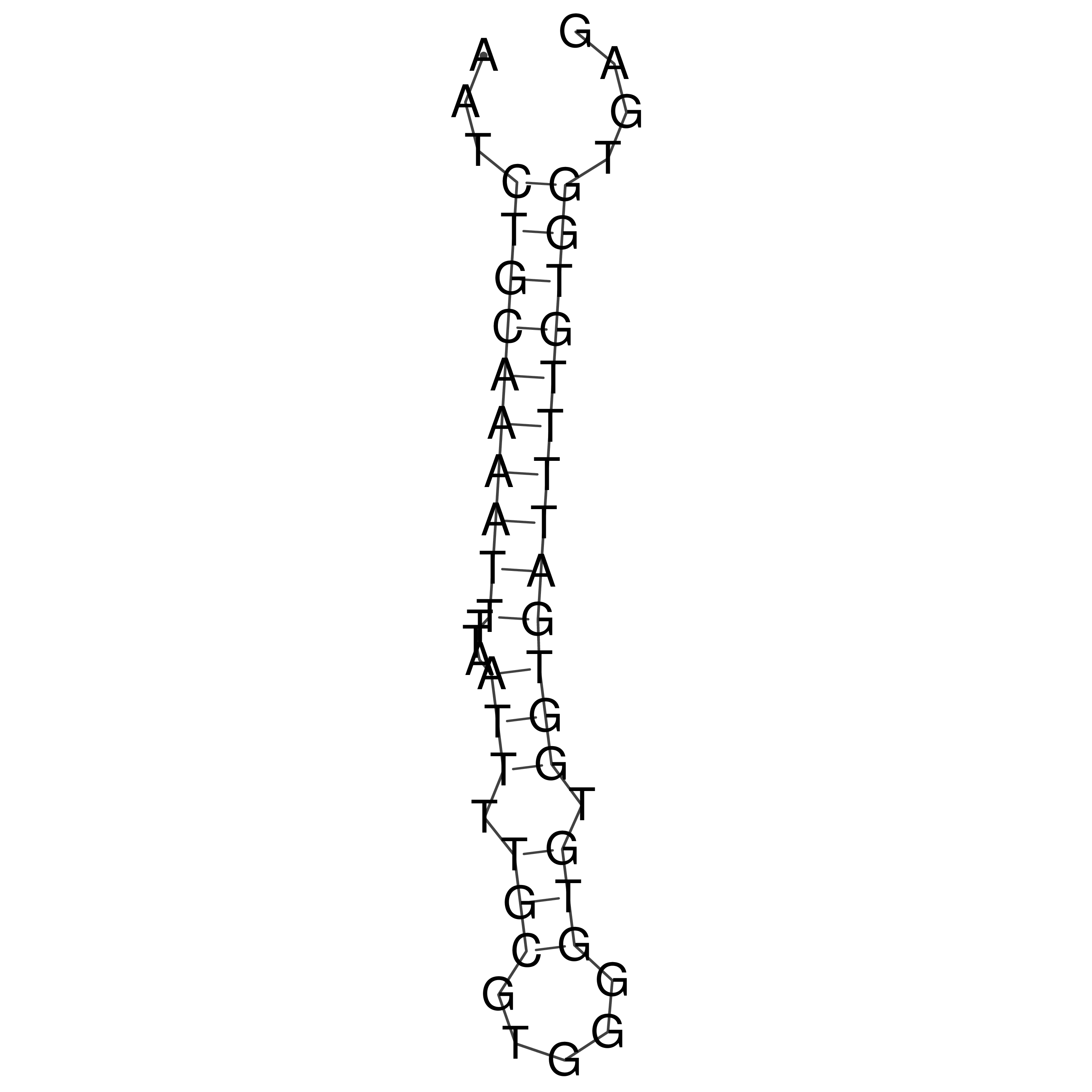
Relaxase
| ID | 10454 | GenBank | WP_064291677 |
| Name | traI_R1019_RS22355_pK118-KPC |
UniProt ID | A0A6I4Y321 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190676.77 Da Isoelectric Point: 6.3386
>WP_064291677.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKDAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQLRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKDAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQLRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12207 | GenBank | WP_072101984 |
| Name | traD_R1019_RS22360_pK118-KPC |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85967.95 Da Isoelectric Point: 5.0577
>WP_072101984.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIS
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPASGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLTDEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIS
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPASGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLTDEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 48678..77151
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| R1019_RS22360 (R1019_22360) | 48678..50990 | - | 2313 | WP_072101984 | type IV conjugative transfer system coupling protein TraD | virb4 |
| R1019_RS22365 (R1019_22365) | 51119..51808 | - | 690 | WP_072101983 | hypothetical protein | - |
| R1019_RS22370 (R1019_22370) | 52001..52732 | - | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| R1019_RS22375 (R1019_22375) | 52918..53451 | - | 534 | WP_014343486 | conjugal transfer protein TraS | - |
| R1019_RS22380 (R1019_22380) | 53454..56303 | - | 2850 | WP_013609534 | conjugal transfer mating-pair stabilization protein TraG | traG |
| R1019_RS22385 (R1019_22385) | 56303..57682 | - | 1380 | WP_014343487 | conjugal transfer pilus assembly protein TraH | traH |
| R1019_RS22390 (R1019_22390) | 57660..58103 | - | 444 | WP_004194305 | F-type conjugal transfer protein TrbF | - |
| R1019_RS22395 (R1019_22395) | 58149..58706 | - | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| R1019_RS22400 (R1019_22400) | 58678..58917 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| R1019_RS22405 (R1019_22405) | 58928..59680 | - | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| R1019_RS22410 (R1019_22410) | 59701..60027 | - | 327 | WP_004152676 | hypothetical protein | - |
| R1019_RS22415 (R1019_22415) | 60040..60288 | - | 249 | WP_004152675 | hypothetical protein | - |
| R1019_RS22420 (R1019_22420) | 60266..60520 | - | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| R1019_RS22425 (R1019_22425) | 60552..62507 | - | 1956 | WP_030003482 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| R1019_RS22430 (R1019_22430) | 62566..63204 | - | 639 | WP_069985712 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| R1019_RS22435 (R1019_22435) | 63217..64206 | - | 990 | WP_160208050 | conjugal transfer pilus assembly protein TraU | traU |
| R1019_RS22440 (R1019_22440) | 64203..64604 | - | 402 | WP_004194979 | hypothetical protein | - |
| R1019_RS22445 (R1019_22445) | 64639..65274 | - | 636 | WP_029497419 | type-F conjugative transfer system protein TraW | traW |
| R1019_RS22450 (R1019_22450) | 65274..65663 | - | 390 | WP_004167468 | type-F conjugative transfer system protein TrbI | - |
| R1019_RS22455 (R1019_22455) | 65663..68302 | - | 2640 | WP_013214024 | type IV secretion system protein TraC | virb4 |
| R1019_RS22460 (R1019_22460) | 68374..68772 | - | 399 | WP_023179972 | hypothetical protein | - |
| R1019_RS22465 (R1019_22465) | 69151..69555 | - | 405 | WP_004197817 | hypothetical protein | - |
| R1019_RS22470 (R1019_22470) | 69622..69933 | - | 312 | WP_029497417 | hypothetical protein | - |
| R1019_RS22475 (R1019_22475) | 69934..70152 | - | 219 | WP_004195468 | hypothetical protein | - |
| R1019_RS22480 (R1019_22480) | 70475..71059 | - | 585 | WP_032440547 | type IV conjugative transfer system lipoprotein TraV | traV |
| R1019_RS22485 (R1019_22485) | 71132..72556 | - | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| R1019_RS22490 (R1019_22490) | 72556..73296 | - | 741 | WP_013023821 | type-F conjugative transfer system secretin TraK | traK |
| R1019_RS22495 (R1019_22495) | 73283..73849 | - | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| R1019_RS22500 (R1019_22500) | 73869..74174 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| R1019_RS22505 (R1019_22505) | 74188..74556 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| R1019_RS22510 (R1019_22510) | 74625..74825 | - | 201 | WP_004194116 | TraY domain-containing protein | - |
| R1019_RS22515 (R1019_22515) | 74911..75612 | - | 702 | WP_004194113 | hypothetical protein | - |
| R1019_RS22520 (R1019_22520) | 75842..76234 | - | 393 | WP_004194114 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| R1019_RS22525 (R1019_22525) | 76666..77151 | + | 486 | WP_032440470 | transglycosylase SLT domain-containing protein | virB1 |
| R1019_RS22530 (R1019_22530) | 77184..77513 | - | 330 | WP_071994324 | DUF5983 family protein | - |
| R1019_RS22535 (R1019_22535) | 77546..78367 | - | 822 | WP_023287138 | DUF932 domain-containing protein | - |
| R1019_RS22540 (R1019_22540) | 79197..79610 | - | 414 | WP_069985713 | type II toxin-antitoxin system HigA family antitoxin | - |
| R1019_RS22545 (R1019_22545) | 79611..79889 | - | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| R1019_RS22550 (R1019_22550) | 79879..80199 | - | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| R1019_RS22555 (R1019_22555) | 80280..80504 | - | 225 | WP_014343499 | hypothetical protein | - |
| R1019_RS22560 (R1019_22560) | 80515..80727 | - | 213 | WP_019706020 | hypothetical protein | - |
| R1019_RS22565 (R1019_22565) | 80788..81144 | - | 357 | WP_019706019 | hypothetical protein | - |
| R1019_RS22570 (R1019_22570) | 81776..82126 | - | 351 | WP_023287104 | hypothetical protein | - |
Host bacterium
| ID | 16797 | GenBank | NZ_CP136818 |
| Plasmid name | pK118-KPC | Incompatibility group | - |
| Plasmid size | 103175 bp | Coordinate of oriT [Strand] | 76545..76593 [+] |
| Host baterium | Citrobacter koseri strain K118 |
Cargo genes
| Drug resistance gene | blaKPC-2, qnrS1 |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |