Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116343 |
| Name | oriT_ICMP 8742|unnamed |
| Organism | Xylella fastidiosa subsp. fastidiosa strain ICMP 8742 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP117824 (20257..20362 [-], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 68..74, 77..83 (CACCGGA..TCCGGTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 106 nt
>oriT_ICMP 8742|unnamed
GCTAGCCTCGCAGAGCAGGATTCCCGTTGAGTGCCGAAGGTGCGAATAAGGGAAAGTGAAGAAGGAACACCGGATTTCCGGTGGGCCTACTTCACATATCCTGCCC
GCTAGCCTCGCAGAGCAGGATTCCCGTTGAGTGCCGAAGGTGCGAATAAGGGAAAGTGAAGAAGGAACACCGGATTTCCGGTGGGCCTACTTCACATATCCTGCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file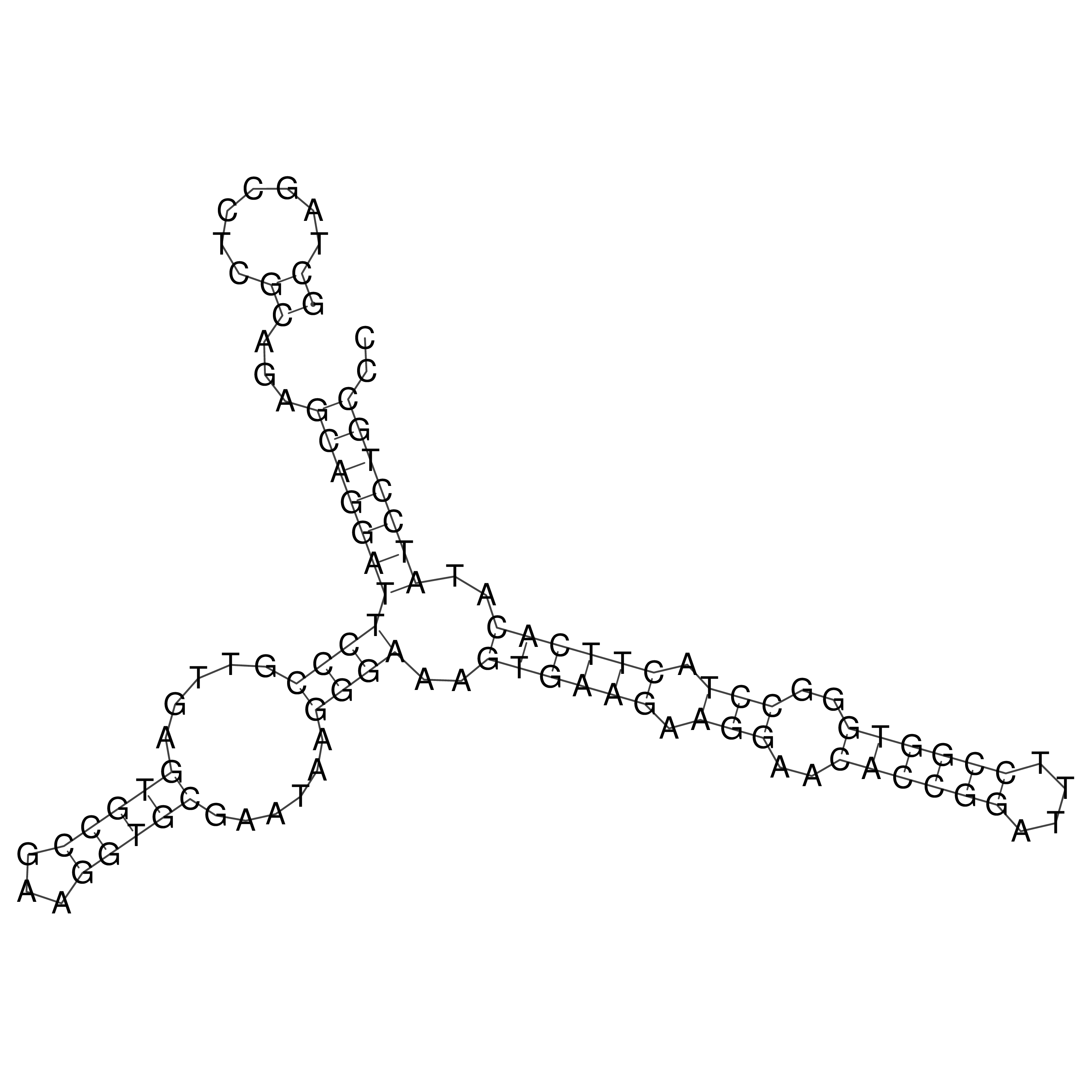
Relaxase
| ID | 10445 | GenBank | WP_004091435 |
| Name | traI_PUO95_RS11695_ICMP 8742|unnamed |
UniProt ID | _ |
| Length | 830 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 830 a.a. Molecular weight: 92723.79 Da Isoelectric Point: 10.6526
>WP_004091435.1 TraI/MobA(P) family conjugative relaxase [Xylella fastidiosa]
MIAKHVPMRVVKKSDFRELVKYLSNQQGKQERVGCVTVTNCFQNNVLDAALEVQATQALNTRSEADKTYH
LLISFREGENPAPEILEVIESRVCAALGYADYQRVSVVHHDTDNLHIHVAINKIHPKRYTIHTPYNDYKT
LGEICKKLEREYGLEADNHTVRKTVGENRADDMERNAGVESLIGWIKRECTDKIKQAQSWSELHAVMQRN
GLEIRERGNGLVITNGVGRSVKASSVARDFSKAKLEARFGAFESFVKQAPSVSSVHARRIQAPLVEKVGR
RPPPRSKGRTPSLSSVGVLHVNSGIRYEQRPIYVKHASTAELYAMYKLEQQDLGAVRNVAIARARTKRDR
KIEATKRLGRIKRAAIKLMRGPGVNKKLLYALARKSLKEGVQKANTDYLKARDAAYATYHRRVWADWLQV
QATQGNAEALAALRAREMRQWWSCNIFGGLQARRTGPVPGLKPDNITKSGTIIYRVGSTAIRDDGDLLNV
SHGAGDYGVEAALRMAMYRYGERITVKGSDEFKKRVVQIAAAARLNISFEDEVLDKRRKQLVSDAAVIVK
LTDAMLSDKTAQFTQAQASRVGAFQEDALSEYDAAISRSINLANQEQINEQQRTRAKHYFVQRWSDRGGP
NSRRDGHATRIRSGGGGSGKHGASGATVISSAGGFSKPNVGGIGAKPPPASTHRLRNLHQLGVVQFSRGS
EVLLPRHVSDCVGEQQTERNHRVRRDVHTAGITSSELMAVDKYIAERELKRVRIFDIMKHRRYNEDDAGM
VKFAGLRQVDGKSLVLLKRNDEVIVLAIDVATARRMKRFSLGDEVLLTKKGTLKTKGRNR
MIAKHVPMRVVKKSDFRELVKYLSNQQGKQERVGCVTVTNCFQNNVLDAALEVQATQALNTRSEADKTYH
LLISFREGENPAPEILEVIESRVCAALGYADYQRVSVVHHDTDNLHIHVAINKIHPKRYTIHTPYNDYKT
LGEICKKLEREYGLEADNHTVRKTVGENRADDMERNAGVESLIGWIKRECTDKIKQAQSWSELHAVMQRN
GLEIRERGNGLVITNGVGRSVKASSVARDFSKAKLEARFGAFESFVKQAPSVSSVHARRIQAPLVEKVGR
RPPPRSKGRTPSLSSVGVLHVNSGIRYEQRPIYVKHASTAELYAMYKLEQQDLGAVRNVAIARARTKRDR
KIEATKRLGRIKRAAIKLMRGPGVNKKLLYALARKSLKEGVQKANTDYLKARDAAYATYHRRVWADWLQV
QATQGNAEALAALRAREMRQWWSCNIFGGLQARRTGPVPGLKPDNITKSGTIIYRVGSTAIRDDGDLLNV
SHGAGDYGVEAALRMAMYRYGERITVKGSDEFKKRVVQIAAAARLNISFEDEVLDKRRKQLVSDAAVIVK
LTDAMLSDKTAQFTQAQASRVGAFQEDALSEYDAAISRSINLANQEQINEQQRTRAKHYFVQRWSDRGGP
NSRRDGHATRIRSGGGGSGKHGASGATVISSAGGFSKPNVGGIGAKPPPASTHRLRNLHQLGVVQFSRGS
EVLLPRHVSDCVGEQQTERNHRVRRDVHTAGITSSELMAVDKYIAERELKRVRIFDIMKHRRYNEDDAGM
VKFAGLRQVDGKSLVLLKRNDEVIVLAIDVATARRMKRFSLGDEVLLTKKGTLKTKGRNR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12196 | GenBank | WP_072866396 |
| Name | t4cp2_PUO95_RS11595_ICMP 8742|unnamed |
UniProt ID | _ |
| Length | 854 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 854 a.a. Molecular weight: 96016.04 Da Isoelectric Point: 6.1729
>WP_072866396.1 conjugal transfer protein TrbE [Xylella fastidiosa]
MFQIITIAIALLGTVLLIWLFLLVQRANADLRLKNHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMASWL
YRGADNASATEAEREFVSFRINQALASLDNAWMIHVDAVRRPAPSYSDPSLSHFPDAVSAAVDEERRRLF
ESIDTLYEGFFVVTVTWFPPVLAERRFVELMFHDDAEKPNDKTRTVHLIDHFKREVINLENRLSLAVELE
RLRGVKITKEDGSTVTHDQQLQWLHFCVTGLNHPIQLPSNPMYIDSLIGGQELYPGVVPKIGRNFIQVVA
IEGFPLESYPGILSVLAELPIEYRWSSRFIFLDHHEAVSHLEKFRKKWKQKMRGFFDQVFNTNTSHVDED
AVSMVADASSAIAETNSGLVSQGYYTSVVVLMNESREQVEHSARRLEKAINALAFTARVETVNTMDAYLG
SLPGHGVENVRRPLLNTMNLADLLPTSTIWTGENRAPSPLFPPNAPPLLHGVTSGNSPMRINLHVRDLGH
CIIFGPTRTGKSTKLALVAFQWRRYLGARIYAFDKGLSMYPTCKAMHGHHFSVASNTDKLAFAPLSRLDT
RTRRAWAMEWIDTILALNGVITSPAQRNAIAEAILSMSESKASTLSEFLVTVQDEVIREALEPYTIDRNM
GYLLDAAEDGLELADFMTFEIEQLMGLGEKFALPVLLYLFMRIEESLSEEDARPTLLILDEAWLMLAHTT
FRDKIEEWLRSMAKKNCSVLMATQSISEAAKSGILDIITESTACRIFLANPNAREEKTAEIYGRLGLNNR
QIEIIASAVPKRDYYYVSEKGRRLYQLALGPLALAFVGSTDKESIAMIRKLEALHGEAWVHEWLRLKGLR
LDEYVSPNDYGVAA
MFQIITIAIALLGTVLLIWLFLLVQRANADLRLKNHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMASWL
YRGADNASATEAEREFVSFRINQALASLDNAWMIHVDAVRRPAPSYSDPSLSHFPDAVSAAVDEERRRLF
ESIDTLYEGFFVVTVTWFPPVLAERRFVELMFHDDAEKPNDKTRTVHLIDHFKREVINLENRLSLAVELE
RLRGVKITKEDGSTVTHDQQLQWLHFCVTGLNHPIQLPSNPMYIDSLIGGQELYPGVVPKIGRNFIQVVA
IEGFPLESYPGILSVLAELPIEYRWSSRFIFLDHHEAVSHLEKFRKKWKQKMRGFFDQVFNTNTSHVDED
AVSMVADASSAIAETNSGLVSQGYYTSVVVLMNESREQVEHSARRLEKAINALAFTARVETVNTMDAYLG
SLPGHGVENVRRPLLNTMNLADLLPTSTIWTGENRAPSPLFPPNAPPLLHGVTSGNSPMRINLHVRDLGH
CIIFGPTRTGKSTKLALVAFQWRRYLGARIYAFDKGLSMYPTCKAMHGHHFSVASNTDKLAFAPLSRLDT
RTRRAWAMEWIDTILALNGVITSPAQRNAIAEAILSMSESKASTLSEFLVTVQDEVIREALEPYTIDRNM
GYLLDAAEDGLELADFMTFEIEQLMGLGEKFALPVLLYLFMRIEESLSEEDARPTLLILDEAWLMLAHTT
FRDKIEEWLRSMAKKNCSVLMATQSISEAAKSGILDIITESTACRIFLANPNAREEKTAEIYGRLGLNNR
QIEIIASAVPKRDYYYVSEKGRRLYQLALGPLALAFVGSTDKESIAMIRKLEALHGEAWVHEWLRLKGLR
LDEYVSPNDYGVAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 12197 | GenBank | WP_004091433 |
| Name | t4cp2_PUO95_RS11700_ICMP 8742|unnamed |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 72064.60 Da Isoelectric Point: 8.5739
>WP_004091433.1 type IV secretory system conjugative DNA transfer family protein [Xylella fastidiosa]
MRKPNNAVGPQVRGGQRKKNKLVPALALTSMATGLQVATQYFAHTFNYQSTLGGNYNHIYAPWSILSWYA
KWGDQFHNYFFAAGNIGMMVTAGGLIGTALIKTFEINSVKANEYLHGSACWASKKDIEQSGLLGNDEGVY
VGAWEDKDGRLHYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWPHSVVVTDLKGELWAMTAGWRQKHAK
NKVIRFEPATLNGSVCWNPLDEIRIGTEYEVGDVQNLATLIVDPNGKGLESHWQKTSQALLVGVILHVLY
KRKNEGFPATLSYVDHIMSDPNRNVADLWMEMTQYGHINGENHPAVGSAGRDMMDRPEEEAGSVLSTAKS
YIALYRDPVVARNVSKSDFCIKDLMNHDDPVSLYIVTQPNDKARLRPLVRVMLNMIVRLLADKMKFERVP
TELSWWRQILTKLGFNQVADFHVRSKKSYKHRLLGMIDEFPSLGKLEIMQESLAFVAGYGIKFYLICQDL
NQLKSRETGYGPDETITSNCHVQNAYPPNRVETAEHLSKLTGQTTVLKEQITTSGKRATPILGQLSRTIQ
EVQRPLLTVDECLRMPGPKKDAEGQITEAGDMVVYVAGYPAIYGKQPLYFQDPIFIARAAVPEPKTSDTL
RLVHEPTLLDEIKI
MRKPNNAVGPQVRGGQRKKNKLVPALALTSMATGLQVATQYFAHTFNYQSTLGGNYNHIYAPWSILSWYA
KWGDQFHNYFFAAGNIGMMVTAGGLIGTALIKTFEINSVKANEYLHGSACWASKKDIEQSGLLGNDEGVY
VGAWEDKDGRLHYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWPHSVVVTDLKGELWAMTAGWRQKHAK
NKVIRFEPATLNGSVCWNPLDEIRIGTEYEVGDVQNLATLIVDPNGKGLESHWQKTSQALLVGVILHVLY
KRKNEGFPATLSYVDHIMSDPNRNVADLWMEMTQYGHINGENHPAVGSAGRDMMDRPEEEAGSVLSTAKS
YIALYRDPVVARNVSKSDFCIKDLMNHDDPVSLYIVTQPNDKARLRPLVRVMLNMIVRLLADKMKFERVP
TELSWWRQILTKLGFNQVADFHVRSKKSYKHRLLGMIDEFPSLGKLEIMQESLAFVAGYGIKFYLICQDL
NQLKSRETGYGPDETITSNCHVQNAYPPNRVETAEHLSKLTGQTTVLKEQITTSGKRATPILGQLSRTIQ
EVQRPLLTVDECLRMPGPKKDAEGQITEAGDMVVYVAGYPAIYGKQPLYFQDPIFIARAAVPEPKTSDTL
RLVHEPTLLDEIKI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1455..10003
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PUO95_RS11565 (PUO95_11565) | 1128..1442 | - | 315 | WP_152534368 | EexN family lipoprotein | - |
| PUO95_RS11570 (PUO95_11570) | 1455..2240 | - | 786 | WP_004091516 | P-type conjugative transfer protein TrbJ | virB5 |
| PUO95_RS11575 (PUO95_11575) | 2259..3659 | - | 1401 | WP_004091515 | TrbI/VirB10 family protein | virB10 |
| PUO95_RS11580 (PUO95_11580) | 3665..4141 | - | 477 | WP_050812601 | conjugal transfer protein TrbH | - |
| PUO95_RS11585 (PUO95_11585) | 4144..5034 | - | 891 | WP_038229217 | P-type conjugative transfer protein TrbG | virB9 |
| PUO95_RS11590 (PUO95_11590) | 5051..5767 | - | 717 | WP_004091508 | conjugal transfer protein TrbF | virB8 |
| PUO95_RS11595 (PUO95_11595) | 5764..8328 | - | 2565 | WP_072866396 | conjugal transfer protein TrbE | virb4 |
| PUO95_RS11600 (PUO95_11600) | 8316..8636 | - | 321 | WP_004091502 | conjugal transfer protein TrbD | virB3 |
| PUO95_RS11605 (PUO95_11605) | 8639..9031 | - | 393 | WP_004091500 | TrbC/VirB2 family protein | virB2 |
| PUO95_RS11610 (PUO95_11610) | 9044..10003 | - | 960 | WP_038232472 | P-type conjugative transfer ATPase TrbB | virB11 |
| PUO95_RS11615 (PUO95_11615) | 10201..10557 | - | 357 | WP_038232465 | transcriptional regulator | - |
| PUO95_RS11620 (PUO95_11620) | 10675..11034 | + | 360 | WP_004091494 | single-stranded DNA-binding protein | - |
| PUO95_RS11625 (PUO95_11625) | 11078..11425 | + | 348 | WP_050812600 | hypothetical protein | - |
| PUO95_RS11630 (PUO95_11630) | 11469..12380 | + | 912 | WP_004091489 | replication protein RepA | - |
| PUO95_RS11635 (PUO95_11635) | 13099..13419 | + | 321 | WP_004091487 | KleE stable inheritance protein | - |
| PUO95_RS11640 (PUO95_11640) | 13615..13914 | + | 300 | WP_038232470 | transcriptional regulator KorA | - |
| PUO95_RS11645 (PUO95_11645) | 13917..14681 | + | 765 | WP_004091486 | ParA family protein | - |
Host bacterium
| ID | 16776 | GenBank | NZ_CP117824 |
| Plasmid name | ICMP 8742|unnamed | Incompatibility group | - |
| Plasmid size | 41752 bp | Coordinate of oriT [Strand] | 20257..20362 [-] |
| Host baterium | Xylella fastidiosa subsp. fastidiosa strain ICMP 8742 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |