Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116341 |
| Name | oriT_H184|unnamed |
| Organism | Butyricimonas faecalis strain H184 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP032820 (47800..47960 [-], 161 nt) |
| oriT length | 161 nt |
| IRs (inverted repeats) | 55..61, 64..70 (CAATGGC..GCCATTG) 16..21, 35..40 (AACTAC..GTAGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 161 nt
>oriT_H184|unnamed
CTCGGGAGAGCCCACAACTACGTAAGCGAAGCGTGTAGTTATAGTGGGCTATATCAATGGCAAGCCATTGTCCGTACACTACAACCTTCGGTTTCCGCTCTCCTCCGTTAAGGAGGGGTTCCGTCACCGCTGCCGATTGAGGATGCACCGACCAGCACAAG
CTCGGGAGAGCCCACAACTACGTAAGCGAAGCGTGTAGTTATAGTGGGCTATATCAATGGCAAGCCATTGTCCGTACACTACAACCTTCGGTTTCCGCTCTCCTCCGTTAAGGAGGGGTTCCGTCACCGCTGCCGATTGAGGATGCACCGACCAGCACAAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file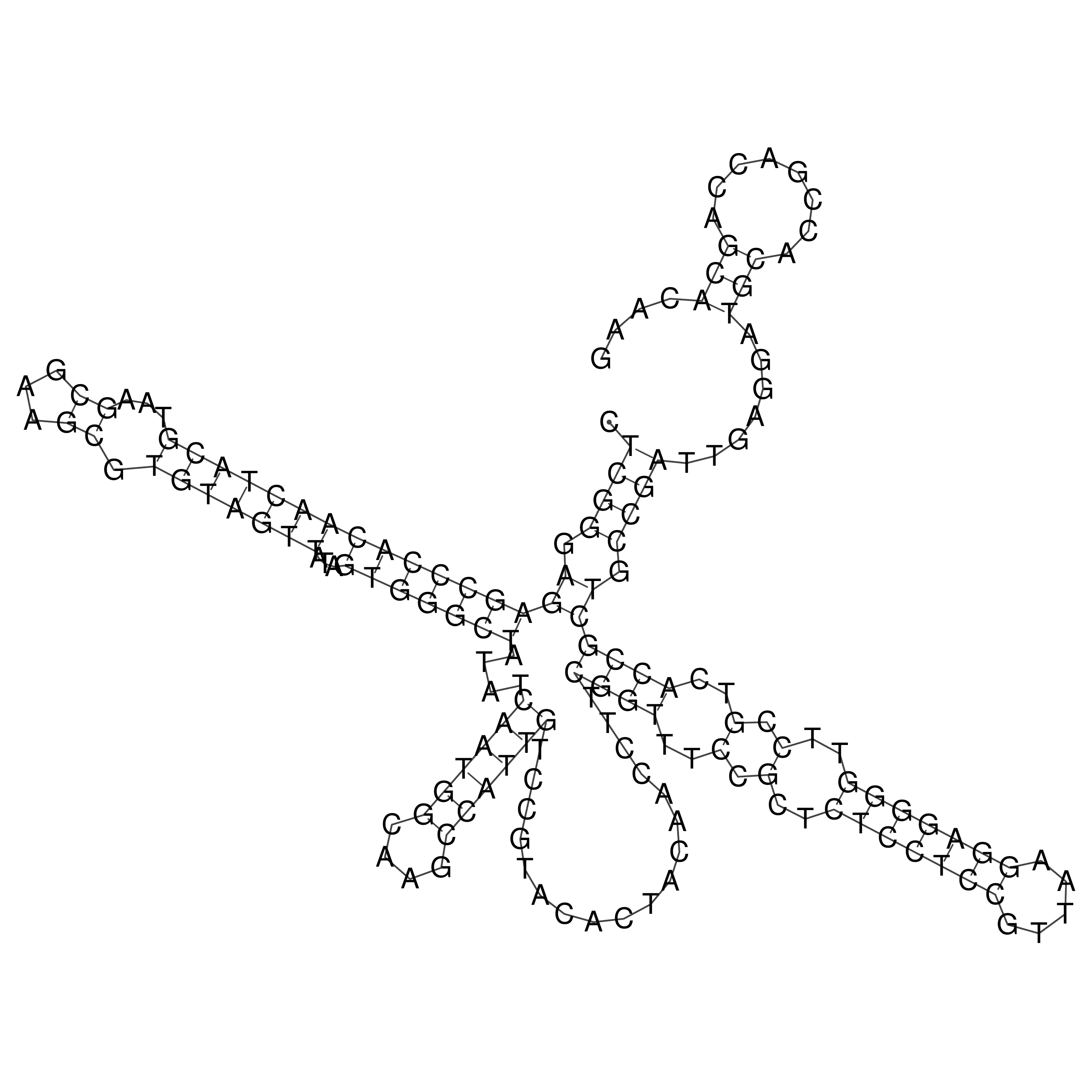
Relaxase
| ID | 10443 | GenBank | WP_007559083 |
| Name | Relaxase_D8S85_RS21175_H184|unnamed |
UniProt ID | A0A3L7ZJB6 |
| Length | 520 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 520 a.a. Molecular weight: 60001.99 Da Isoelectric Point: 9.4388
>WP_007559083.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Bacteria]
MVVVVLKAATSFAGIDYNERKNEEGKSELLVAENFAMNPDNLKKSDYIAYMESVCRTNPAVKAKQFHAVI
SCKGREYSAEDLKNVALQYINKMGYGNNPYMIYFHSDTENNHVHIVSTRVQKNGQKVKDNMEAVRSQKVI
NQIMNVDLALKAKDDISKYMEFSFSTVQQYKLLLEQSGWKLREKDGLLILYKGGEKHASIQLEQIKDKAK
QYTPDEERRKQITALLFKYKQGLSYTELQTLMKDKFGINLVFHTGKGHTKPYGYTLIDYRNKRVLKGGEV
MDLKELLVEPNKQAKVEHCNSIINLLLKDGTKYTMDSFKKLMLDYGYRFSMNGTIFINGDDKALLVLDKK
LLKELRYNSRVYEANKFNVATLKEAELLSRIYHVKKDDILIQEHMDKKNTVYTDMINSYLAHSSDLHSTL
REKGILFVEDDNLVYLIDKKNKVIVSTDDLGINIIKDGYSKDRITLVTLDKMERINVEQDSELARGFNLI
DAICDILSQHINVQQDKSPNRKKKRGQQQN
MVVVVLKAATSFAGIDYNERKNEEGKSELLVAENFAMNPDNLKKSDYIAYMESVCRTNPAVKAKQFHAVI
SCKGREYSAEDLKNVALQYINKMGYGNNPYMIYFHSDTENNHVHIVSTRVQKNGQKVKDNMEAVRSQKVI
NQIMNVDLALKAKDDISKYMEFSFSTVQQYKLLLEQSGWKLREKDGLLILYKGGEKHASIQLEQIKDKAK
QYTPDEERRKQITALLFKYKQGLSYTELQTLMKDKFGINLVFHTGKGHTKPYGYTLIDYRNKRVLKGGEV
MDLKELLVEPNKQAKVEHCNSIINLLLKDGTKYTMDSFKKLMLDYGYRFSMNGTIFINGDDKALLVLDKK
LLKELRYNSRVYEANKFNVATLKEAELLSRIYHVKKDDILIQEHMDKKNTVYTDMINSYLAHSSDLHSTL
REKGILFVEDDNLVYLIDKKNKVIVSTDDLGINIIKDGYSKDRITLVTLDKMERINVEQDSELARGFNLI
DAICDILSQHINVQQDKSPNRKKKRGQQQN
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3L7ZJB6 |
T4CP
| ID | 12195 | GenBank | WP_004295345 |
| Name | t4cp2_D8S85_RS21150_H184|unnamed |
UniProt ID | _ |
| Length | 651 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 651 a.a. Molecular weight: 74504.94 Da Isoelectric Point: 8.6510
>WP_004295345.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Bacteria]
MEESKELQKLYGFMQAFIYITVCIEILLFVHFPFSEQIMPLLSKMAKIPIYSNILYSKLFTFFIIMVTCI
GTRSKKDLELDPTKQIIFPLIFGFIMFFGSICFLFFKEKGETSLEWYNIAYIILSIIGATLINSALDNIS
KRIKSNFMKDRFNIENESFEQSKDKVETEFSVNIPMKFFYNRKWHNGWLNICNCFRGTFVIGTPGSGKSF
SVINSFIRQHSAKGFAEVVYDFKFPELAKIAYYNYQKNKQLGKIPSNFKFNVINFSDIEYSRRINPLKRE
YIEILADATETAEALYESLQKGDKGSGGNSDFFKTSAVNLLAASIYFWSRYENGKYSDLPHVLAFLNQEY
DVLFKVLFSEPELKSLVSPFEAAYKSGAVDQLEGQMASLKVQLSRLATKESFWVFSGNDFNLKVSDKKDP
SYLIIANNPKTQSMNSALNALIINRLTRLVNTKGNYPTSIIVDECPTLYFYQLATLLSTARSNKVSICLG
LQELPQLEEQYGKATAKTITSIIGNTLSGQAKAPETLDWLQKLFGKVKQVKEGVTIRRNETTINMNEQMD
FVIPASKISSLQAGTLVGQVALDFGQEDNFPTAMYHCKTNLDLKKIKKEEEAYKELPKVYNFGTADNREK
LLQKNFKRIYDEVETVIEQYV
MEESKELQKLYGFMQAFIYITVCIEILLFVHFPFSEQIMPLLSKMAKIPIYSNILYSKLFTFFIIMVTCI
GTRSKKDLELDPTKQIIFPLIFGFIMFFGSICFLFFKEKGETSLEWYNIAYIILSIIGATLINSALDNIS
KRIKSNFMKDRFNIENESFEQSKDKVETEFSVNIPMKFFYNRKWHNGWLNICNCFRGTFVIGTPGSGKSF
SVINSFIRQHSAKGFAEVVYDFKFPELAKIAYYNYQKNKQLGKIPSNFKFNVINFSDIEYSRRINPLKRE
YIEILADATETAEALYESLQKGDKGSGGNSDFFKTSAVNLLAASIYFWSRYENGKYSDLPHVLAFLNQEY
DVLFKVLFSEPELKSLVSPFEAAYKSGAVDQLEGQMASLKVQLSRLATKESFWVFSGNDFNLKVSDKKDP
SYLIIANNPKTQSMNSALNALIINRLTRLVNTKGNYPTSIIVDECPTLYFYQLATLLSTARSNKVSICLG
LQELPQLEEQYGKATAKTITSIIGNTLSGQAKAPETLDWLQKLFGKVKQVKEGVTIRRNETTINMNEQMD
FVIPASKISSLQAGTLVGQVALDFGQEDNFPTAMYHCKTNLDLKKIKKEEEAYKELPKVYNFGTADNREK
LLQKNFKRIYDEVETVIEQYV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 16774 | GenBank | NZ_CP032820 |
| Plasmid name | H184|unnamed | Incompatibility group | - |
| Plasmid size | 51017 bp | Coordinate of oriT [Strand] | 47800..47960 [-] |
| Host baterium | Butyricimonas faecalis strain H184 |
Cargo genes
| Drug resistance gene | cfxA |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |