Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116312 |
| Name | oriT_pAt2 |
| Organism | Agrobacterium tumefaciens strain Sut001 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP048517 (103331..103371 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pAt2
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file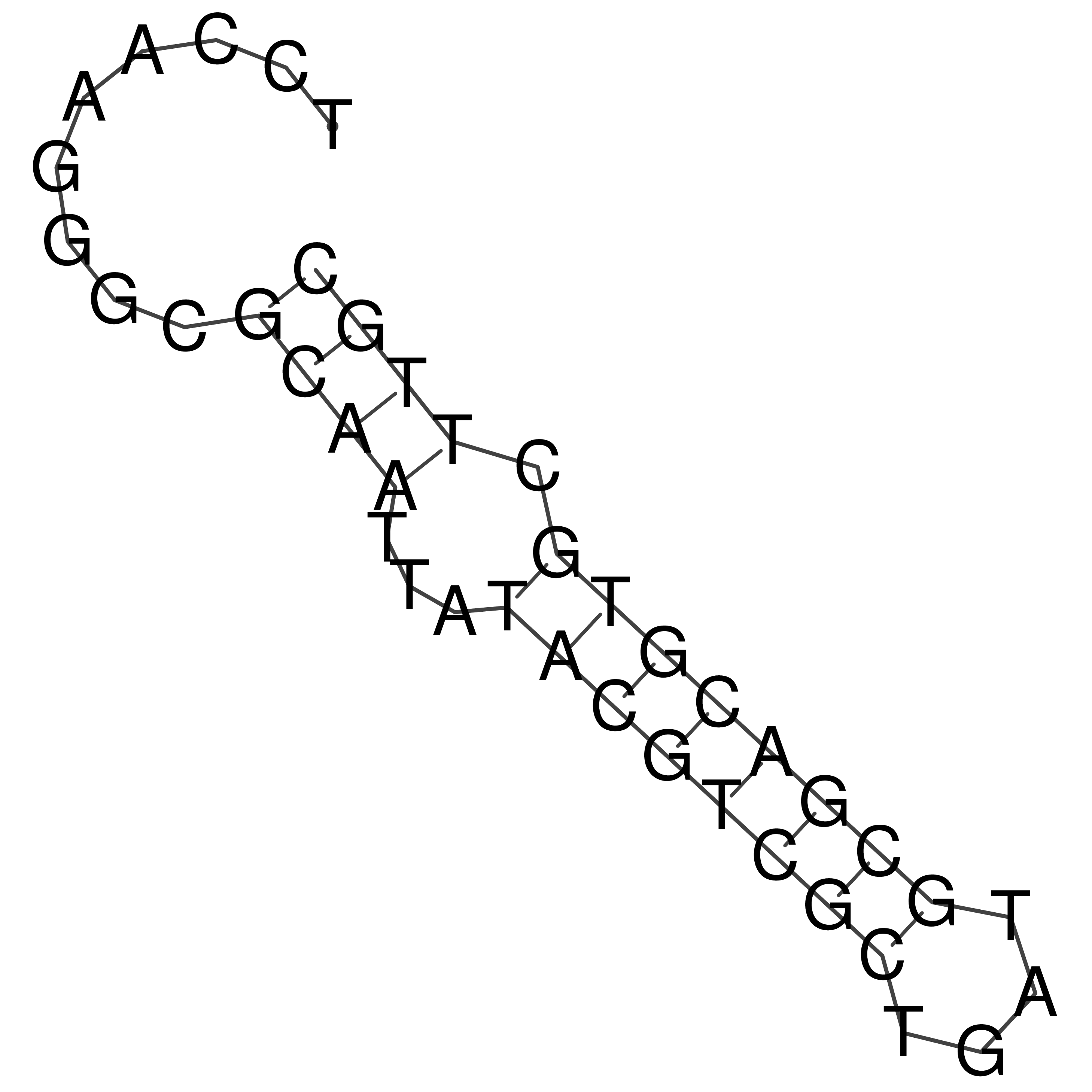
Relaxase
| ID | 10423 | GenBank | WP_173995559 |
| Name | traA_FY145_RS28100_pAt2 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123269.13 Da Isoelectric Point: 9.9132
>WP_173995559.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium/Agrobacterium group]
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLVHEEFMLPADAPKWVRAMIADR
SVAGASEAFWNKVEAFEKRADAQLARDLTIALPLELPAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTGDGFGTKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIDLEPTIHLGVGAKAIERKAKEQGIRPELERIELNEDRRSENARRILRRPEIVLDL
ITREKSVFDERDVAKVLHRYVDDPALFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMARQAMWLSDKETHAVSTAVLAATFGRHGRLSEEQKAAIECIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIHSRTLASWELRWNRGRDVLDRKTVFVMDEAGMVASKQMARFVD
TAVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNVMAREKFVERGVVGEGHVFKTADG
IRQFDAGDQIVFLKNETSLGVKNGMIAHVVEAAPNRIVAVVGEGDKRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDVQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAEGRGLNIVQVARTLVRDRLDWTLRQKMKLVELGQRLAVFAEHLGLHHPLKTQTMKEAAPMVAG
IKTFSGSVADTLSDRLGADPALKQQWEEVSARFAYVFADPETAFRAMNFDAVLADKDSARQALQKLEAEP
TSIGALTGKTGILASKTEREARRIAELNVPALKRDLEQYLRMRETASQRLQADEQALRQRVSIDIPALSP
AACVVLERVRDAIDRNDLPAALGYALSNRETKLEIDGFNKAVTERFGERTLLTNAARQPSGTLFDKLSNG
MQPAQKEQLREAWPVMRTAQQIAAHERTTQTLKQAEELRLTQRQSPVMKQ
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLVHEEFMLPADAPKWVRAMIADR
SVAGASEAFWNKVEAFEKRADAQLARDLTIALPLELPAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTGDGFGTKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIDLEPTIHLGVGAKAIERKAKEQGIRPELERIELNEDRRSENARRILRRPEIVLDL
ITREKSVFDERDVAKVLHRYVDDPALFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMARQAMWLSDKETHAVSTAVLAATFGRHGRLSEEQKAAIECIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIHSRTLASWELRWNRGRDVLDRKTVFVMDEAGMVASKQMARFVD
TAVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNVMAREKFVERGVVGEGHVFKTADG
IRQFDAGDQIVFLKNETSLGVKNGMIAHVVEAAPNRIVAVVGEGDKRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDVQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAEGRGLNIVQVARTLVRDRLDWTLRQKMKLVELGQRLAVFAEHLGLHHPLKTQTMKEAAPMVAG
IKTFSGSVADTLSDRLGADPALKQQWEEVSARFAYVFADPETAFRAMNFDAVLADKDSARQALQKLEAEP
TSIGALTGKTGILASKTEREARRIAELNVPALKRDLEQYLRMRETASQRLQADEQALRQRVSIDIPALSP
AACVVLERVRDAIDRNDLPAALGYALSNRETKLEIDGFNKAVTERFGERTLLTNAARQPSGTLFDKLSNG
MQPAQKEQLREAWPVMRTAQQIAAHERTTQTLKQAEELRLTQRQSPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12169 | GenBank | WP_173995561 |
| Name | traG_FY145_RS28085_pAt2 |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 71160.44 Da Isoelectric Point: 9.0431
>WP_173995561.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MTMNRLLLLILPAIIMLAAMFATSGMEQRLAAFGTSPRAKLMLGHAGLALPYIAAVAIGIIGLFAANGSA
NIKAAGLSVLAGNGVVITTATLREVIRLNSIASRVPAEQSVLAYADPVTMIGVGIAFISGMFALRVAIKG
NAAFSTTAPKRIGGKRAVHGEADWMKIQETAKLFPEPGGIVIGERYRVDRDSVAAMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVCEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEA
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGNVEGRTLFLLDEVARLGY
LRILETARDAGRKYGIMLTMIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
ENRFHRTGTGTDTHTEHAPPWQKEGTRP
MTMNRLLLLILPAIIMLAAMFATSGMEQRLAAFGTSPRAKLMLGHAGLALPYIAAVAIGIIGLFAANGSA
NIKAAGLSVLAGNGVVITTATLREVIRLNSIASRVPAEQSVLAYADPVTMIGVGIAFISGMFALRVAIKG
NAAFSTTAPKRIGGKRAVHGEADWMKIQETAKLFPEPGGIVIGERYRVDRDSVAAMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVCEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEA
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGNVEGRTLFLLDEVARLGY
LRILETARDAGRKYGIMLTMIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
ENRFHRTGTGTDTHTEHAPPWQKEGTRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 12170 | GenBank | WP_173995645 |
| Name | t4cp2_FY145_RS28410_pAt2 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 92049.83 Da Isoelectric Point: 5.5826
>WP_173995645.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium/Agrobacterium group]
MVALKLFRHSGPSFADLVPYAGLVDSGIILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINLILSRLG
SGWMIQVEALRVPTIDYPAGDICHFPDPVTRAIDTERRSHFERESGHFESRHALILTWRPPEPRRSGLTR
YVYSDTASRSATYADTALDTFRTSIREVEQYLANVLSIRRMVTRETEERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVL
FDEQQERLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLSLIAAQFRRYADAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDAAEGGEGDSPRLAFCPLAELASDGDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGAFQCFEVEEL
MNLGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRNKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHFEHGADWPKHWLQQRGIAHAETLFQAA
MVALKLFRHSGPSFADLVPYAGLVDSGIILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINLILSRLG
SGWMIQVEALRVPTIDYPAGDICHFPDPVTRAIDTERRSHFERESGHFESRHALILTWRPPEPRRSGLTR
YVYSDTASRSATYADTALDTFRTSIREVEQYLANVLSIRRMVTRETEERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVL
FDEQQERLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLSLIAAQFRRYADAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDAAEGGEGDSPRLAFCPLAELASDGDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGAFQCFEVEEL
MNLGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRNKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHFEHGADWPKHWLQQRGIAHAETLFQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 164392..174064
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FY145_RS28350 (FY145_28165) | 159676..160401 | - | 726 | WP_173995637 | nopaline ABC transporter permease NocM | - |
| FY145_RS28355 (FY145_28170) | 160403..161113 | - | 711 | WP_173995654 | nopaline ABC transporter permease NocQ | - |
| FY145_RS28360 (FY145_28175) | 161187..162038 | - | 852 | WP_173995638 | nopaline ABC transporter substrate-binding protein NocT | - |
| FY145_RS28365 (FY145_28180) | 162103..162876 | - | 774 | WP_173995639 | nopaline ABC transporter ATP-binding protein NocP | - |
| FY145_RS28370 (FY145_28185) | 163130..164032 | + | 903 | WP_045023359 | LysR family transcriptional regulator | - |
| FY145_RS28375 (FY145_28190) | 164392..165699 | - | 1308 | WP_174168418 | IncP-type conjugal transfer protein TrbI | virB10 |
| FY145_RS28380 (FY145_28195) | 165714..166190 | - | 477 | WP_045025162 | conjugal transfer protein TrbH | - |
| FY145_RS28385 (FY145_28200) | 166190..167044 | - | 855 | WP_173995641 | P-type conjugative transfer protein TrbG | virB9 |
| FY145_RS28390 (FY145_28205) | 167062..167724 | - | 663 | WP_060716494 | conjugal transfer protein TrbF | virB8 |
| FY145_RS28395 (FY145_28210) | 167739..168935 | - | 1197 | WP_173995642 | P-type conjugative transfer protein TrbL | virB6 |
| FY145_RS28400 (FY145_28215) | 168929..169147 | - | 219 | WP_173995643 | entry exclusion protein TrbK | - |
| FY145_RS28405 (FY145_28220) | 169144..169908 | - | 765 | Protein_153 | P-type conjugative transfer protein TrbJ | - |
| FY145_RS28410 (FY145_28225) | 169925..172393 | - | 2469 | WP_173995645 | conjugal transfer protein TrbE | virb4 |
| FY145_RS28415 (FY145_28230) | 172404..172703 | - | 300 | WP_173995646 | conjugal transfer protein TrbD | virB3 |
| FY145_RS28420 (FY145_28235) | 172696..173103 | - | 408 | WP_173995647 | conjugal transfer pilin TrbC | virB2 |
| FY145_RS28425 (FY145_28240) | 173093..174064 | - | 972 | WP_173995648 | P-type conjugative transfer ATPase TrbB | virB11 |
| FY145_RS28430 (FY145_28245) | 174061..174696 | - | 636 | WP_173995649 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 16745 | GenBank | NZ_CP048517 |
| Plasmid name | pAt2 | Incompatibility group | - |
| Plasmid size | 175061 bp | Coordinate of oriT [Strand] | 103331..103371 [+] |
| Host baterium | Agrobacterium tumefaciens strain Sut001 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |